| schema location: | http://bioxsd.org/BioXSD-1.1.xsd |
| attribute form default: | unqualified |
| element form default: | qualified |
| targetNamespace: | http://bioxsd.org/BioXSD-1.1 |
GeneralNucleotideSequence simpleType
| type | restriction of xs:string | |||||||||
| used by |
|
|||||||||
| facets |
|
|||||||||
| documentation | Nucleotide sequence, possibly with ambiguous ("degenerate") bases | |||||||||
| source | <xs:simpleType name="GeneralNucleotideSequence" sawsdl:modelReference="http://edamontology.org/data_2975 http://edamontology.org/format_1210 http://edamontology.org/format_1957"> <xs:annotation> <xs:documentation>Nucleotide sequence, possibly with ambiguous ("degenerate") bases</xs:documentation> </xs:annotation> <xs:restriction base="xs:string"> <xs:pattern value="[acgtrykmswbdhvn]+"/> <xs:pattern value="[acgurykmswbdhvn]+"/> </xs:restriction> </xs:simpleType> |
GeneralAminoacidSequence simpleType
| type | restriction of xs:string | |||||||||
| used by |
|
|||||||||
| facets |
|
|||||||||
| documentation | Amino-acid sequence, possibly with ambiguous residues (Asx, Xle, Glx, Xaa/Unk) and additional residues (Pyl and Sec) | |||||||||
| source | <xs:simpleType name="GeneralAminoacidSequence" sawsdl:modelReference="http://edamontology.org/data_2974 http://edamontology.org/format_1219 http://edamontology.org/format_1957"> <xs:annotation> <xs:documentation>Amino-acid sequence, possibly with ambiguous residues (Asx, Xle, Glx, Xaa/Unk) and additional residues (Pyl and Sec)</xs:documentation> </xs:annotation> <xs:restriction base="xs:string"> <xs:maxLength value="1000000000"/> <xs:pattern value="[ACDEFGHIKLMNPQRSTVWYBJZXUO]+"/> </xs:restriction> </xs:simpleType> |
Biosequence simpleType
| type | union of (GeneralNucleotideSequence, GeneralAminoacidSequence) | ||
| used by |
|
||
| documentation | Nucleotide or amino-acid sequence, possibly with ambiguous bases and residues | ||
| source | <xs:simpleType name="Biosequence" sawsdl:modelReference="http://edamontology.org/data_0848 http://edamontology.org/format_2094 http://edamontology.org/format_1957"> <xs:annotation> <xs:documentation>Nucleotide or amino-acid sequence, possibly with ambiguous bases and residues</xs:documentation> </xs:annotation> <xs:union memberTypes="GeneralNucleotideSequence GeneralAminoacidSequence"/> </xs:simpleType> |
NucleotideSequence simpleType
| type | restriction of GeneralNucleotideSequence | |||||||||
| used by |
|
|||||||||
| facets |
|
|||||||||
| documentation | Nucleotide sequence without ambiguous ("degenerate") bases | |||||||||
| source | <xs:simpleType name="NucleotideSequence" sawsdl:modelReference="http://edamontology.org/format_2568"> <xs:annotation> <xs:documentation>Nucleotide sequence without ambiguous ("degenerate") bases</xs:documentation> </xs:annotation> <xs:restriction base="GeneralNucleotideSequence"> <xs:pattern value="[acgt]+"/> <xs:pattern value="[acgu]+"/> </xs:restriction> </xs:simpleType> |
AminoacidSequence simpleType
| type | restriction of GeneralAminoacidSequence | |||||||||
| used by |
|
|||||||||
| facets |
|
|||||||||
| documentation | Amino-acid sequence without ambiguous and additional residues (Pyl, Sec) | |||||||||
| source | <xs:simpleType name="AminoacidSequence" sawsdl:modelReference="http://edamontology.org/format_2607"> <xs:annotation> <xs:documentation>Amino-acid sequence without ambiguous and additional residues (Pyl, Sec)</xs:documentation> </xs:annotation> <xs:restriction base="GeneralAminoacidSequence"> <xs:pattern value="[ACDEFGHIKLMNPQRSTVWY]+"/> </xs:restriction> </xs:simpleType> |
BaseQualityString simpleType
| type | restriction of xs:string | ||||||
| used by |
|
||||||
| facets |
|
||||||
| source | <xs:simpleType name="BaseQualityString"> <xs:restriction base="xs:string"> <xs:pattern value="[!-~]+"/> </xs:restriction> </xs:simpleType> |
BiosequenceRecord complexType
| diagram | 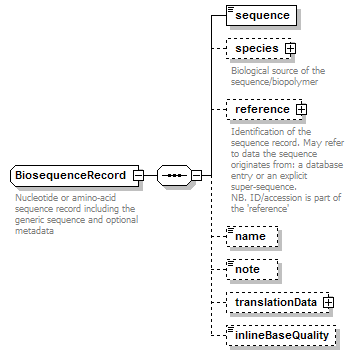 |
||||
| children | sequence species reference name note translationData inlineBaseQuality | ||||
| used by |
|
||||
| documentation | Nucleotide or amino-acid sequence record including the generic sequence and optional metadata | ||||
| source | <xs:complexType name="BiosequenceRecord" sawsdl:modelReference="http://edamontology.org/data_2043 http://edamontology.org/format_2352"> <xs:annotation> <xs:documentation>Nucleotide or amino-acid sequence record including the generic sequence and optional metadata</xs:documentation> </xs:annotation> <xs:sequence> <xs:element name="sequence" type="Biosequence"/> <xs:element name="species" type="Species" minOccurs="0"> <xs:annotation> <xs:documentation>Biological source of the sequence/biopolymer</xs:documentation> </xs:annotation> </xs:element> <xs:element name="reference" type="SequenceReference" minOccurs="0"> <xs:annotation> <xs:documentation>Identification of the sequence record. May refer to data the sequence originates from: a database entry or an explicit super-sequence. NB. ID/accession is part of the 'reference'</xs:documentation> </xs:annotation> </xs:element> <xs:element name="name" type="Name" minOccurs="0" sawsdl:modelReference="http://www.w3.org/2000/01/rdf-schema#label http://edamontology.org/data_2154"/> <!--"Sequence name"(2154)--> <xs:element name="note" type="Text" minOccurs="0" sawsdl:modelReference="http://www.w3.org/2000/01/rdf-schema#comment"/> <xs:element name="translationData" type="TranslationData" minOccurs="0"/> <xs:element name="inlineBaseQuality" type="BaseQualityString" minOccurs="0"/> </xs:sequence> </xs:complexType> |
||||
| model reference | http://edamontology.org/data_2043 http://edamontology.org/format_2352 |
GeneralNucleotideSequenceRecord complexType
| diagram | 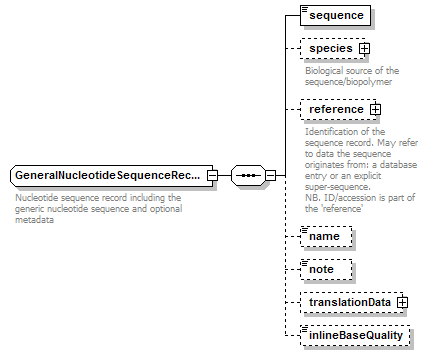 |
||||
| type | restriction of BiosequenceRecord | ||||
| properties |
|
||||
| children | sequence species reference name note translationData inlineBaseQuality | ||||
| used by |
|
||||
| documentation | Nucleotide sequence record including the generic nucleotide sequence and optional metadata | ||||
| source | <xs:complexType name="GeneralNucleotideSequenceRecord" sawsdl:modelReference="http://edamontology.org/data_2046 http://edamontology.org/format_2352"> <xs:annotation> <xs:documentation>Nucleotide sequence record including the generic nucleotide sequence and optional metadata</xs:documentation> </xs:annotation> <xs:complexContent> <xs:restriction base="BiosequenceRecord"> <xs:sequence> <xs:element name="sequence" type="GeneralNucleotideSequence"/> <xs:element name="species" type="Species" minOccurs="0"> <xs:annotation> <xs:documentation>Biological source of the sequence/biopolymer</xs:documentation> </xs:annotation> </xs:element> <xs:element name="reference" type="SequenceReference" minOccurs="0"> <xs:annotation> <xs:documentation>Identification of the sequence record. May refer to data the sequence originates from: a database entry or an explicit super-sequence. NB. ID/accession is part of the 'reference'</xs:documentation> </xs:annotation> </xs:element> <xs:element name="name" type="Name" minOccurs="0"/> <xs:element name="note" type="Text" minOccurs="0"/> <xs:element name="translationData" type="TranslationData" minOccurs="0"/> <xs:element name="inlineBaseQuality" type="BaseQualityString" minOccurs="0"/> </xs:sequence> </xs:restriction> </xs:complexContent> </xs:complexType> |
||||
| model reference | http://edamontology.org/data_2046 http://edamontology.org/format_2352 |
GeneralAminoacidSequenceRecord complexType
| diagram |  |
||||
| type | restriction of BiosequenceRecord | ||||
| properties |
|
||||
| children | sequence species reference name note translationData | ||||
| used by |
|
||||
| documentation | Amino-acid sequence record including the generic amino-acid sequence and optional metadata | ||||
| source | <xs:complexType name="GeneralAminoacidSequenceRecord" sawsdl:modelReference="http://edamontology.org/data_2047 http://edamontology.org/format_2352"> <xs:annotation> <xs:documentation>Amino-acid sequence record including the generic amino-acid sequence and optional metadata</xs:documentation> </xs:annotation> <xs:complexContent> <xs:restriction base="BiosequenceRecord"> <xs:sequence> <xs:element name="sequence" type="GeneralAminoacidSequence"/> <xs:element name="species" type="Species" minOccurs="0"> <xs:annotation> <xs:documentation>Biological source of the sequence/biopolymer</xs:documentation> </xs:annotation> </xs:element> <xs:element name="reference" type="SequenceReference" minOccurs="0"> <xs:annotation> <xs:documentation>Identification of the sequence record. May refer to data the sequence originates from: a database entry or an explicit super-sequence. NB. ID/accession is part of the 'reference'</xs:documentation> </xs:annotation> </xs:element> <xs:element name="name" type="Name" minOccurs="0"/> <xs:element name="note" type="Text" minOccurs="0"/> <xs:element name="translationData" type="AminoacidTranslationData" minOccurs="0"/> </xs:sequence> </xs:restriction> </xs:complexContent> </xs:complexType> |
||||
| model reference | http://edamontology.org/data_2047 http://edamontology.org/format_2352 |
NucleotideSequenceRecord complexType
| diagram | 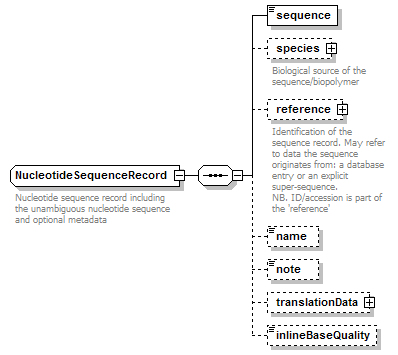 |
||
| type | restriction of GeneralNucleotideSequenceRecord | ||
| properties |
|
||
| children | sequence species reference name note translationData inlineBaseQuality | ||
| used by |
|
||
| documentation | Nucleotide sequence record including the unambiguous nucleotide sequence and optional metadata | ||
| source | <xs:complexType name="NucleotideSequenceRecord" sawsdl:modelReference="http://edamontology.org/data_2046 http://edamontology.org/format_2352"> <xs:annotation> <xs:documentation>Nucleotide sequence record including the unambiguous nucleotide sequence and optional metadata</xs:documentation> </xs:annotation> <xs:complexContent> <xs:restriction base="GeneralNucleotideSequenceRecord"> <xs:sequence> <xs:element name="sequence" type="NucleotideSequence"/> <xs:element name="species" type="Species" minOccurs="0"> <xs:annotation> <xs:documentation>Biological source of the sequence/biopolymer</xs:documentation> </xs:annotation> </xs:element> <xs:element name="reference" type="SequenceReference" minOccurs="0"> <xs:annotation> <xs:documentation>Identification of the sequence record. May refer to data the sequence originates from: a database entry or an explicit super-sequence. NB. ID/accession is part of the 'reference'</xs:documentation> </xs:annotation> </xs:element> <xs:element name="name" type="Name" minOccurs="0"/> <xs:element name="note" type="Text" minOccurs="0"/> <xs:element name="translationData" type="TranslationData" minOccurs="0"/> <xs:element name="inlineBaseQuality" type="BaseQualityString" minOccurs="0"/> </xs:sequence> </xs:restriction> </xs:complexContent> </xs:complexType> |
||
| model reference | http://edamontology.org/data_2046 http://edamontology.org/format_2352 |
AminoacidSequenceRecord complexType
| diagram | 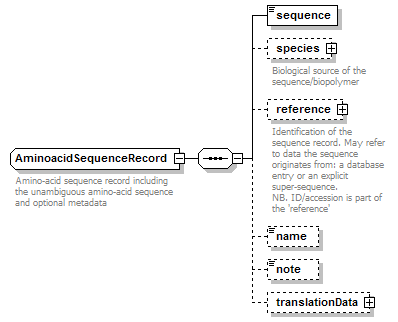 |
||
| type | restriction of GeneralAminoacidSequenceRecord | ||
| properties |
|
||
| children | sequence species reference name note translationData | ||
| used by |
|
||
| documentation | Amino-acid sequence record including the unambiguous amino-acid sequence and optional metadata | ||
| source | <xs:complexType name="AminoacidSequenceRecord" sawsdl:modelReference="http://edamontology.org/data_2047 http://edamontology.org/format_2352"> <xs:annotation> <xs:documentation>Amino-acid sequence record including the unambiguous amino-acid sequence and optional metadata</xs:documentation> </xs:annotation> <xs:complexContent> <xs:restriction base="GeneralAminoacidSequenceRecord"> <xs:sequence> <xs:element name="sequence" type="AminoacidSequence"/> <xs:element name="species" type="Species" minOccurs="0"> <xs:annotation> <xs:documentation>Biological source of the sequence/biopolymer</xs:documentation> </xs:annotation> </xs:element> <xs:element name="reference" type="SequenceReference" minOccurs="0"> <xs:annotation> <xs:documentation>Identification of the sequence record. May refer to data the sequence originates from: a database entry or an explicit super-sequence. NB. ID/accession is part of the 'reference'</xs:documentation> </xs:annotation> </xs:element> <xs:element name="name" type="Name" minOccurs="0"/> <xs:element name="note" type="Text" minOccurs="0"/> <xs:element name="translationData" type="AminoacidTranslationData" minOccurs="0"/> </xs:sequence> </xs:restriction> </xs:complexContent> </xs:complexType> |
||
| model reference | http://edamontology.org/data_2047 http://edamontology.org/format_2352 |
BiosequenceAlignment complexType
| diagram | 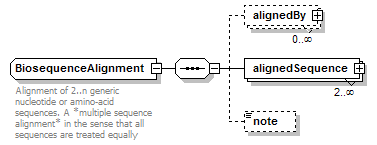 |
||
| children | alignedBy alignedSequence note | ||
| used by |
|
||
| documentation | Alignment of 2..n generic nucleotide or amino-acid sequences. A *multiple sequence alignment* in the sense that all sequences are treated equally | ||
| source | <xs:complexType name="BiosequenceAlignment" sawsdl:modelReference="http://edamontology.org/data_0863 http://edamontology.org/format_2352"> <xs:annotation> <xs:documentation>Alignment of 2..n generic nucleotide or amino-acid sequences. A *multiple sequence alignment* in the sense that all sequences are treated equally</xs:documentation> </xs:annotation> <xs:sequence> <xs:element name="alignedBy" type="MethodResult" minOccurs="0" maxOccurs="unbounded"/> <xs:element name="alignedSequence" type="AlignedBiosequence" minOccurs="2" maxOccurs="unbounded"> <xs:unique name="uniqueGapPos_b"> <xs:selector xpath="gap"/> <xs:field xpath="@pos"/> </xs:unique> <xs:unique name="uniqueInsertPos_b"> <xs:selector xpath="insert"/> <xs:field xpath="@min"/> <xs:field xpath="@max"/> </xs:unique> <xs:unique name="uniqueFrameshiftPos_b"> <xs:selector xpath="frameshift"/> <xs:field xpath="@pos"/> </xs:unique> </xs:element> <xs:element name="note" type="Text" minOccurs="0" sawsdl:modelReference="http://www.w3.org/2000/01/rdf-schema#comment"/> </xs:sequence> </xs:complexType> |
||
| model reference | http://edamontology.org/data_0863 http://edamontology.org/format_2352 |
GeneralNucleotideSequenceAlignment complexType
| diagram | 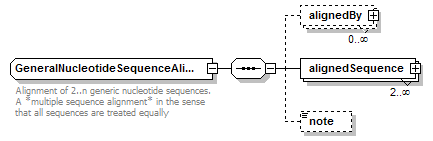 |
||
| type | restriction of BiosequenceAlignment | ||
| properties |
|
||
| children | alignedBy alignedSequence note | ||
| used by |
|
||
| documentation | Alignment of 2..n generic nucleotide sequences. A *multiple sequence alignment* in the sense that all sequences are treated equally | ||
| source | <xs:complexType name="GeneralNucleotideSequenceAlignment" sawsdl:modelReference="http://edamontology.org/data_1383"> <xs:annotation> <xs:documentation>Alignment of 2..n generic nucleotide sequences. A *multiple sequence alignment* in the sense that all sequences are treated equally</xs:documentation> </xs:annotation> <xs:complexContent> <xs:restriction base="BiosequenceAlignment"> <xs:sequence> <xs:element name="alignedBy" type="MethodResult" minOccurs="0" maxOccurs="unbounded"/> <xs:element name="alignedSequence" type="AlignedGeneralNucleotideSequence" minOccurs="2" maxOccurs="unbounded"/> <xs:element name="note" type="Text" minOccurs="0"/> </xs:sequence> </xs:restriction> </xs:complexContent> </xs:complexType> |
||
| model reference | http://edamontology.org/data_1383 |
GeneralAminoacidSequenceAlignment complexType
| diagram | 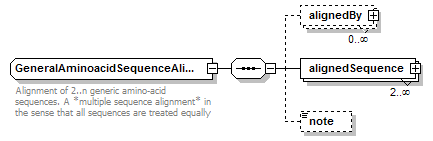 |
||
| type | restriction of BiosequenceAlignment | ||
| properties |
|
||
| children | alignedBy alignedSequence note | ||
| used by |
|
||
| documentation | Alignment of 2..n generic amino-acid sequences. A *multiple sequence alignment* in the sense that all sequences are treated equally | ||
| source | <xs:complexType name="GeneralAminoacidSequenceAlignment" sawsdl:modelReference="http://edamontology.org/data_1384"> <xs:annotation> <xs:documentation>Alignment of 2..n generic amino-acid sequences. A *multiple sequence alignment* in the sense that all sequences are treated equally</xs:documentation> </xs:annotation> <xs:complexContent> <xs:restriction base="BiosequenceAlignment"> <xs:sequence> <xs:element name="alignedBy" type="MethodResult" minOccurs="0" maxOccurs="unbounded"/> <xs:element name="alignedSequence" type="AlignedGeneralAminoacidSequence" minOccurs="2" maxOccurs="unbounded"/> <xs:element name="note" type="Text" minOccurs="0"/> </xs:sequence> </xs:restriction> </xs:complexContent> </xs:complexType> |
||
| model reference | http://edamontology.org/data_1384 |
NucleotideSequenceAlignment complexType
| diagram | 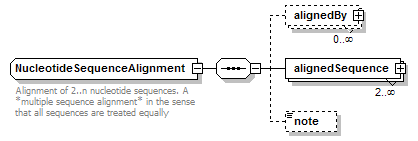 |
||
| type | restriction of GeneralNucleotideSequenceAlignment | ||
| properties |
|
||
| children | alignedBy alignedSequence note | ||
| documentation | Alignment of 2..n nucleotide sequences. A *multiple sequence alignment* in the sense that all sequences are treated equally | ||
| source | <xs:complexType name="NucleotideSequenceAlignment"> <xs:annotation> <xs:documentation>Alignment of 2..n nucleotide sequences. A *multiple sequence alignment* in the sense that all sequences are treated equally</xs:documentation> </xs:annotation> <xs:complexContent> <xs:restriction base="GeneralNucleotideSequenceAlignment"> <xs:sequence> <xs:element name="alignedBy" type="MethodResult" minOccurs="0" maxOccurs="unbounded"/> <xs:element name="alignedSequence" type="AlignedNucleotideSequence" minOccurs="2" maxOccurs="unbounded"/> <xs:element name="note" type="Text" minOccurs="0"/> </xs:sequence> </xs:restriction> </xs:complexContent> </xs:complexType> |
AminoacidSequenceAlignment complexType
| diagram | 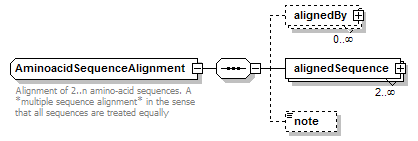 |
||
| type | restriction of GeneralAminoacidSequenceAlignment | ||
| properties |
|
||
| children | alignedBy alignedSequence note | ||
| documentation | Alignment of 2..n amino-acid sequences. A *multiple sequence alignment* in the sense that all sequences are treated equally | ||
| source | <xs:complexType name="AminoacidSequenceAlignment"> <xs:annotation> <xs:documentation>Alignment of 2..n amino-acid sequences. A *multiple sequence alignment* in the sense that all sequences are treated equally</xs:documentation> </xs:annotation> <xs:complexContent> <xs:restriction base="GeneralAminoacidSequenceAlignment"> <xs:sequence> <xs:element name="alignedBy" type="MethodResult" minOccurs="0" maxOccurs="unbounded"/> <xs:element name="alignedSequence" type="AlignedAminoacidSequence" minOccurs="2" maxOccurs="unbounded"/> <xs:element name="note" type="Text" minOccurs="0"/> </xs:sequence> </xs:restriction> </xs:complexContent> </xs:complexType> |
Gap complexType
| diagram | 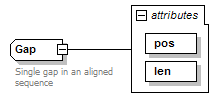 |
||||||||||||||||||
| used by | |||||||||||||||||||
| attributes |
|
||||||||||||||||||
| documentation | Single gap in an aligned sequence | ||||||||||||||||||
| source | <xs:complexType name="Gap"> <xs:annotation> <xs:documentation>Single gap in an aligned sequence</xs:documentation> </xs:annotation> <xs:attribute name="pos" type="InsertionInteger" use="required" sawsdl:modelReference="http://edamontology.org/data_1016"/> <xs:attribute name="len" type="PositiveInteger" use="required"/> </xs:complexType> |
Insert complexType
| diagram | 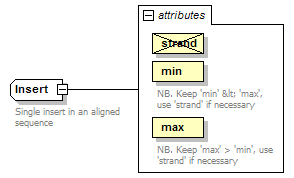 |
||||||||||||||||||||||||||||
| type | restriction of SequenceSegment | ||||||||||||||||||||||||||||
| properties |
|
||||||||||||||||||||||||||||
| used by | |||||||||||||||||||||||||||||
| attributes |
|
||||||||||||||||||||||||||||
| documentation | Single insert in an aligned sequence | ||||||||||||||||||||||||||||
| source | <xs:complexType name="Insert"> <xs:annotation> <xs:documentation>Single insert in an aligned sequence</xs:documentation> </xs:annotation> <xs:complexContent> <xs:restriction base="SequenceSegment"> <xs:attribute name="strand" type="Strand" use="prohibited"/> </xs:restriction> </xs:complexContent> </xs:complexType> |
Frameshift complexType
| diagram | 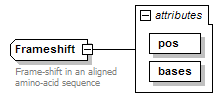 |
||||||||||||||||||
| used by |
|
||||||||||||||||||
| attributes |
|
||||||||||||||||||
| documentation | Frame-shift in an aligned amino-acid sequence | ||||||||||||||||||
| source | <xs:complexType name="Frameshift"> <xs:annotation> <xs:documentation>Frame-shift in an aligned amino-acid sequence</xs:documentation> </xs:annotation> <xs:attribute name="pos" type="InsertionInteger" use="required" sawsdl:modelReference="http://edamontology.org/data_1016"/> <xs:attribute name="bases" use="required"> <xs:simpleType> <xs:restriction base="NonzeroInteger"> <xs:minInclusive value="-2"/> <xs:maxInclusive value="2"/> </xs:restriction> </xs:simpleType> </xs:attribute> </xs:complexType> |
AlignedBiosequence complexType
| diagram | 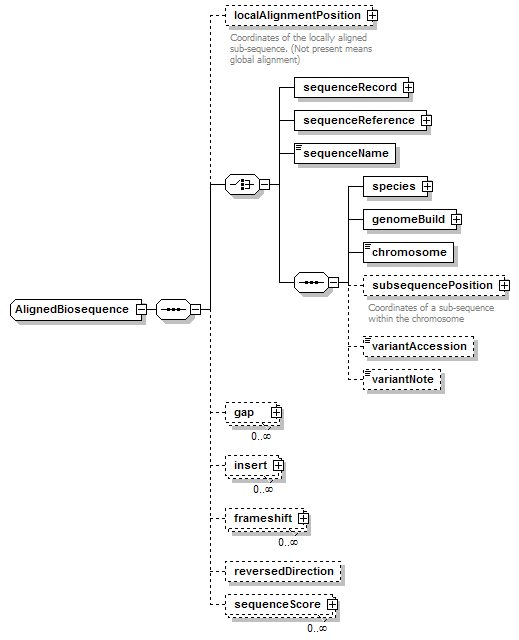 |
||||
| children | localAlignmentPosition sequenceRecord sequenceReference sequenceName species genomeBuild chromosome subsequencePosition variantAccession variantNote gap insert frameshift reversedDirection sequenceScore | ||||
| used by |
|
||||
| source | <xs:complexType name="AlignedBiosequence"> <xs:sequence> <xs:element name="localAlignmentPosition" type="SequencePosition" minOccurs="0"> <xs:annotation> <xs:documentation>Coordinates of the locally aligned sub-sequence. (Not present means global alignment)</xs:documentation> </xs:annotation> </xs:element> <xs:choice> <xs:element name="sequenceRecord" type="BiosequenceRecord"/> <xs:element name="sequenceReference" type="SequenceReference"/> <xs:element name="sequenceName" type="Name" sawsdl:modelReference="http://edamontology.org/data_2154"/> <xs:sequence> <xs:element name="species" type="Species"/> <xs:element name="genomeBuild" type="EntryReference" sawsdl:modelReference="http://purl.org/dc/elements/1.1/version http://edamontology.org/data_2041 http://edamontology.org/data_2340"/> <!--"Genome build identifier"(2340)--> <!--"Genome version information"(2041)--> <xs:element name="chromosome" type="ChromosomeName"/> <xs:element name="subsequencePosition" type="SequencePosition" minOccurs="0"> <xs:annotation> <xs:documentation>Coordinates of a sub-sequence within the chromosome</xs:documentation> </xs:annotation> </xs:element> <xs:element name="variantAccession" type="Accession" minOccurs="0" sawsdl:modelReference="http://edamontology.org/data_1063 http://edamontology.org/data_2091"/> <xs:element name="variantNote" type="Name" minOccurs="0" sawsdl:modelReference="http://edamontology.org/data_1063"/> </xs:sequence> </xs:choice> <xs:element name="gap" type="Gap" minOccurs="0" maxOccurs="unbounded"/> <xs:element name="insert" type="Insert" minOccurs="0" maxOccurs="unbounded"/> <xs:element name="frameshift" type="Frameshift" minOccurs="0" maxOccurs="unbounded"/> <xs:element name="reversedDirection" type="Flag" minOccurs="0"/> <xs:element name="sequenceScore" type="Result" minOccurs="0" maxOccurs="unbounded"/> </xs:sequence> </xs:complexType> |
AlignedGeneralNucleotideSequence complexType
| diagram | 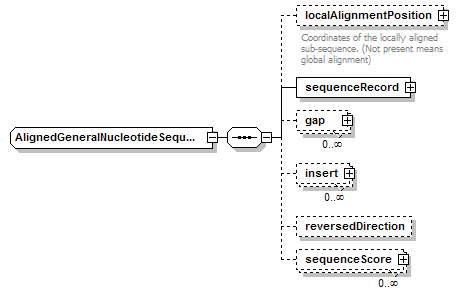 |
||||
| type | restriction of AlignedBiosequence | ||||
| properties |
|
||||
| children | localAlignmentPosition sequenceRecord gap insert reversedDirection sequenceScore | ||||
| used by |
|
||||
| source | <xs:complexType name="AlignedGeneralNucleotideSequence"> <xs:complexContent> <xs:restriction base="AlignedBiosequence"> <xs:sequence> <xs:element name="localAlignmentPosition" type="SequencePosition" minOccurs="0"> <xs:annotation> <xs:documentation>Coordinates of the locally aligned sub-sequence. (Not present means global alignment)</xs:documentation> </xs:annotation> </xs:element> <xs:element name="sequenceRecord" type="GeneralNucleotideSequenceRecord"/> <xs:element name="gap" type="Gap" minOccurs="0" maxOccurs="unbounded"/> <xs:element name="insert" type="Insert" minOccurs="0" maxOccurs="unbounded"/> <xs:element name="reversedDirection" type="Flag" minOccurs="0"/> <xs:element name="sequenceScore" type="Result" minOccurs="0" maxOccurs="unbounded"/> </xs:sequence> </xs:restriction> </xs:complexContent> </xs:complexType> |
AlignedGeneralAminoacidSequence complexType
| diagram | 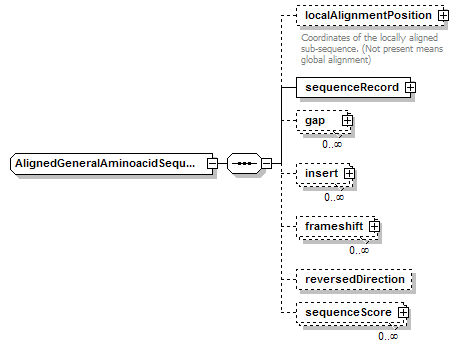 |
||||
| type | restriction of AlignedBiosequence | ||||
| properties |
|
||||
| children | localAlignmentPosition sequenceRecord gap insert frameshift reversedDirection sequenceScore | ||||
| used by |
|
||||
| source | <xs:complexType name="AlignedGeneralAminoacidSequence"> <xs:complexContent> <xs:restriction base="AlignedBiosequence"> <xs:sequence> <xs:element name="localAlignmentPosition" type="SequencePosition" minOccurs="0"> <xs:annotation> <xs:documentation>Coordinates of the locally aligned sub-sequence. (Not present means global alignment)</xs:documentation> </xs:annotation> </xs:element> <xs:element name="sequenceRecord" type="GeneralAminoacidSequenceRecord"/> <xs:element name="gap" type="Gap" minOccurs="0" maxOccurs="unbounded"/> <xs:element name="insert" type="Insert" minOccurs="0" maxOccurs="unbounded"/> <xs:element name="frameshift" type="Frameshift" minOccurs="0" maxOccurs="unbounded"/> <xs:element name="reversedDirection" type="Flag" minOccurs="0"/> <xs:element name="sequenceScore" type="Result" minOccurs="0" maxOccurs="unbounded"/> </xs:sequence> </xs:restriction> </xs:complexContent> </xs:complexType> |
AlignedNucleotideSequence complexType
| diagram | 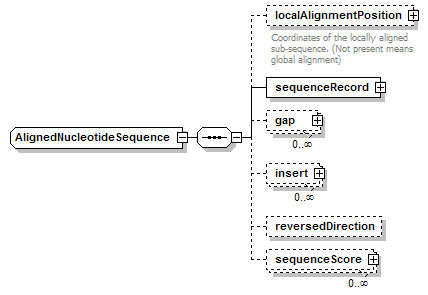 |
||
| type | restriction of AlignedGeneralNucleotideSequence | ||
| properties |
|
||
| children | localAlignmentPosition sequenceRecord gap insert reversedDirection sequenceScore | ||
| used by |
|
||
| source | <xs:complexType name="AlignedNucleotideSequence"> <xs:complexContent> <xs:restriction base="AlignedGeneralNucleotideSequence"> <xs:sequence> <xs:element name="localAlignmentPosition" type="SequencePosition" minOccurs="0"> <xs:annotation> <xs:documentation>Coordinates of the locally aligned sub-sequence. (Not present means global alignment)</xs:documentation> </xs:annotation> </xs:element> <xs:element name="sequenceRecord" type="NucleotideSequenceRecord"/> <xs:element name="gap" type="Gap" minOccurs="0" maxOccurs="unbounded"/> <xs:element name="insert" type="Insert" minOccurs="0" maxOccurs="unbounded"/> <xs:element name="reversedDirection" type="Flag" minOccurs="0"/> <xs:element name="sequenceScore" type="Result" minOccurs="0" maxOccurs="unbounded"/> </xs:sequence> </xs:restriction> </xs:complexContent> </xs:complexType> |
AlignedAminoacidSequence complexType
| diagram |  |
||
| type | restriction of AlignedGeneralAminoacidSequence | ||
| properties |
|
||
| children | localAlignmentPosition sequenceRecord gap insert frameshift reversedDirection sequenceScore | ||
| used by |
|
||
| source | <xs:complexType name="AlignedAminoacidSequence"> <xs:complexContent> <xs:restriction base="AlignedGeneralAminoacidSequence"> <xs:sequence> <xs:element name="localAlignmentPosition" type="SequencePosition" minOccurs="0"> <xs:annotation> <xs:documentation>Coordinates of the locally aligned sub-sequence. (Not present means global alignment)</xs:documentation> </xs:annotation> </xs:element> <xs:element name="sequenceRecord" type="AminoacidSequenceRecord"/> <xs:element name="gap" type="Gap" minOccurs="0" maxOccurs="unbounded"/> <xs:element name="insert" type="Insert" minOccurs="0" maxOccurs="unbounded"/> <xs:element name="frameshift" type="Frameshift" minOccurs="0" maxOccurs="unbounded"/> <xs:element name="reversedDirection" type="Flag" minOccurs="0"/> <xs:element name="sequenceScore" type="Result" minOccurs="0" maxOccurs="unbounded"/> </xs:sequence> </xs:restriction> </xs:complexContent> </xs:complexType> |
FeatureType complexType
| diagram | 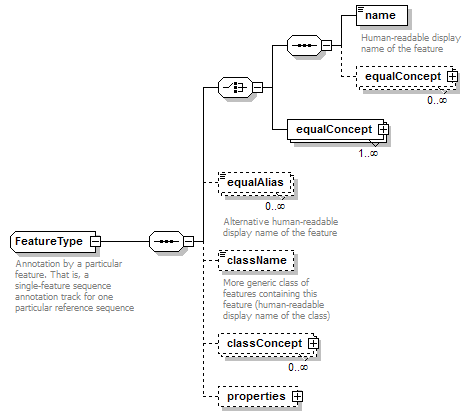 |
||
| children | name equalConcept equalConcept equalAlias className classConcept properties | ||
| used by |
|
||
| documentation | Annotation by a particular feature. That is, a single-feature sequence annotation track for one particular reference sequence | ||
| source | <xs:complexType name="FeatureType" sawsdl:modelReference="http://edamontology.org/data_1015 http://edamontology.org/format_2352"> <xs:annotation> <xs:documentation>Annotation by a particular feature. That is, a single-feature sequence annotation track for one particular reference sequence</xs:documentation> </xs:annotation> <xs:sequence> <xs:choice> <xs:sequence> <xs:element name="name" type="Name" sawsdl:modelReference="http://edamontology.org/data_1015"> <xs:annotation> <xs:documentation>Human-readable display name of the feature</xs:documentation> </xs:annotation> <!--"Sequence feature identifier"(1015)--> </xs:element> <xs:element name="equalConcept" type="SemanticConcept" minOccurs="0" maxOccurs="unbounded" sawsdl:modelReference="http://edamontology.org/data_1015"/> <!--"Sequence feature identifier"(1015)--> </xs:sequence> <xs:element name="equalConcept" type="SemanticConcept" maxOccurs="unbounded" sawsdl:modelReference="http://edamontology.org/data_1015"/> <!--"Sequence feature identifier"(1015)--> </xs:choice> <xs:element name="equalAlias" type="Name" minOccurs="0" maxOccurs="unbounded" sawsdl:modelReference="http://edamontology.org/data_1015"> <xs:annotation> <xs:documentation>Alternative human-readable display name of the feature</xs:documentation> </xs:annotation> <!--"Sequence feature identifier"(1015)--> </xs:element> <xs:element name="className" type="Name" minOccurs="0"> <xs:annotation> <xs:documentation>More generic class of features containing this feature (human-readable display name of the class)</xs:documentation> </xs:annotation> </xs:element> <xs:element name="classConcept" type="SemanticConcept" minOccurs="0" maxOccurs="unbounded"/> <xs:element name="properties" type="FeatureTypeDetails" minOccurs="0"/> </xs:sequence> <!--"Sequence feature identifier"(1015)--> <!--"BioXSD"(2352)--> </xs:complexType> |
||
| model reference | http://edamontology.org/data_1015 http://edamontology.org/format_2352 |
FeatureTypeDetails complexType
| diagram | 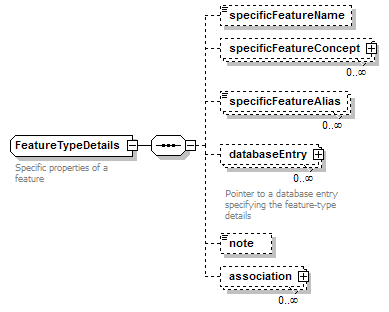 |
||
| children | specificFeatureName specificFeatureConcept specificFeatureAlias databaseEntry note association | ||
| used by |
|
||
| documentation | Specific properties of a feature | ||
| source | <xs:complexType name="FeatureTypeDetails" sawsdl:modelReference="http://edamontology.org/data_1021 http://edamontology.org/format_2352"> <xs:annotation> <xs:documentation>Specific properties of a feature</xs:documentation> </xs:annotation> <xs:sequence> <xs:element name="specificFeatureName" type="Name" minOccurs="0" sawsdl:modelReference="http://edamontology.org/data_1020"/> <!--"Sequence feature qualifier"(1021)--> <xs:element name="specificFeatureConcept" type="SemanticConcept" minOccurs="0" maxOccurs="unbounded"/> <!--"Sequence feature qualifier"(1021)--> <xs:element name="specificFeatureAlias" type="Name" minOccurs="0" maxOccurs="unbounded" sawsdl:modelReference="http://edamontology.org/data_1020"/> <!--"Sequence feature qualifier"(1021)--> <xs:element name="databaseEntry" type="EntryReference" minOccurs="0" maxOccurs="unbounded"> <xs:annotation> <xs:documentation>Pointer to a database entry specifying the feature-type details</xs:documentation> </xs:annotation> </xs:element> <xs:element name="note" type="Text" minOccurs="0" sawsdl:modelReference="http://www.w3.org/2000/01/rdf-schema#comment"/> <xs:element name="association" minOccurs="0" maxOccurs="unbounded" sawsdl:modelReference="http://edamontology.org/data_2093 http://edamontology.org/format_2352"> <xs:complexType> <xs:sequence> <xs:element name="relationName" type="Name" minOccurs="0"/> <xs:element name="relationConcept" type="SemanticConcept" minOccurs="0" maxOccurs="unbounded"/> <xs:choice> <xs:element name="associatedConcept" type="SemanticConcept" maxOccurs="unbounded"/> <xs:element name="associatedDatabaseEntry" type="EntryReference" maxOccurs="unbounded"/> <xs:element name="associatedDatabase" type="DatabaseReference" maxOccurs="unbounded"/> <xs:element name="associatedFeature" type="FeatureType" maxOccurs="unbounded"/> </xs:choice> </xs:sequence> </xs:complexType> </xs:element> </xs:sequence> <!--"Sequence feature qualifier"(1021)--> <!--"BioXSD"(2352)--> </xs:complexType> |
||
| model reference | http://edamontology.org/data_1021 http://edamontology.org/format_2352 |
FeatureOccurrenceData complexType
| diagram | 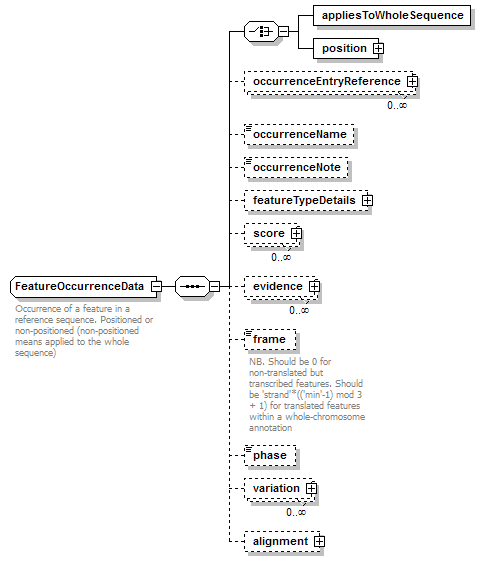 |
||
| children | appliesToWholeSequence position occurrenceEntryReference occurrenceName occurrenceNote featureTypeDetails score evidence frame phase variation alignment | ||
| used by |
|
||
| documentation | Occurrence of a feature in a reference sequence. Positioned or non-positioned (non-positioned means applied to the whole sequence) | ||
| source | <xs:complexType name="FeatureOccurrenceData" sawsdl:modelReference="http://edamontology.org/data_1021 http://edamontology.org/format_2352"> <xs:annotation> <xs:documentation>Occurrence of a feature in a reference sequence. Positioned or non-positioned (non-positioned means applied to the whole sequence)</xs:documentation> </xs:annotation> <xs:sequence> <xs:choice> <xs:element name="appliesToWholeSequence" type="Flag"/> <xs:element name="position" type="GeneralSequencePosition"/> </xs:choice> <xs:element name="occurrenceEntryReference" type="EntryReference" minOccurs="0" maxOccurs="unbounded"/> <xs:element name="occurrenceName" type="Name" minOccurs="0"/> <xs:element name="occurrenceNote" type="Text" minOccurs="0"/> <xs:element name="featureTypeDetails" type="FeatureTypeDetails" minOccurs="0"/> <xs:element name="score" type="Score" minOccurs="0" maxOccurs="unbounded"/> <xs:element name="evidence" type="Evidence" minOccurs="0" maxOccurs="unbounded"/> <xs:element name="frame" minOccurs="0" sawsdl:modelReference="http://edamontology.org/data_2126"> <xs:annotation> <xs:documentation>NB. Should be 0 for non-translated but transcribed features. Should be 'strand'*(('min'-1) mod 3 + 1) for translated features within a whole-chromosome annotation</xs:documentation> </xs:annotation> <xs:simpleType> <xs:restriction base="AnyInteger"> <xs:minInclusive value="-3"/> <xs:maxInclusive value="3"/> </xs:restriction> </xs:simpleType> </xs:element> <xs:element name="phase" type="Phase" minOccurs="0"/> <xs:element name="variation" minOccurs="0" maxOccurs="unbounded" sawsdl:modelReference="http://edamontology.org/data_0918 http://edamontology.org/format_2352"> <xs:complexType> <xs:sequence> <xs:element name="canonicalVariant" type="Biosequence" minOccurs="0"/> <xs:choice> <xs:element name="variant" type="Biosequence" maxOccurs="unbounded"/> <xs:element name="missing" type="Flag"/> </xs:choice> <xs:element name="score" type="Score" minOccurs="0" maxOccurs="unbounded"/> <xs:element name="position" type="SequencePosition" minOccurs="0"> <xs:annotation> <xs:documentation>NB. Corresponds to a position within the feature occurrence</xs:documentation> </xs:annotation> </xs:element> </xs:sequence> </xs:complexType> </xs:element> <xs:element name="alignment" minOccurs="0" sawsdl:modelReference="http://edamontology.org/data_1381 http://edamontology.org/format_2352"> <xs:complexType> <xs:sequence> <xs:element name="alignmentOfReference" minOccurs="0"> <xs:complexType> <xs:sequence> <xs:element name="gap" type="Gap" minOccurs="0" maxOccurs="unbounded"/> <xs:element name="insert" type="Insert" minOccurs="0" maxOccurs="unbounded"/> <xs:element name="frameshift" type="Frameshift" minOccurs="0" maxOccurs="unbounded"/> <xs:element name="sequenceScore" type="Result" minOccurs="0" maxOccurs="unbounded"/> </xs:sequence> </xs:complexType> <xs:unique name="uniqueGapPos_ar"> <xs:selector xpath="gap"/> <xs:field xpath="@pos"/> </xs:unique> <xs:unique name="uniqueInsertPos_ar"> <xs:selector xpath="insert"/> <xs:field xpath="@min"/> <xs:field xpath="@max"/> </xs:unique> <xs:unique name="uniqueFrameshiftPos_ar"> <xs:selector xpath="frameshift"/> <xs:field xpath="@pos"/> </xs:unique> </xs:element> <xs:element name="alignedSequence" type="AlignedBiosequence"> <xs:unique name="uniqueGapPos_as"> <xs:selector xpath="gap"/> <xs:field xpath="@pos"/> </xs:unique> <xs:unique name="uniqueInsertPos_as"> <xs:selector xpath="insert"/> <xs:field xpath="@min"/> <xs:field xpath="@max"/> </xs:unique> <xs:unique name="uniqueFrameshiftPos_as"> <xs:selector xpath="frameshift"/> <xs:field xpath="@pos"/> </xs:unique> </xs:element> </xs:sequence> </xs:complexType> </xs:element> </xs:sequence> </xs:complexType> |
||
| model reference | http://edamontology.org/data_1021 http://edamontology.org/format_2352 |
FeatureRecord complexType
| diagram |  |
| children | referenceSequence annotation blockWithOccurrenceReferences |
| documentation | Features of a sequence/biopolymer (or a genome/metagenome).The reference sequence may me included explicitly, referred to remotely, or implicit |
| source | <xs:complexType name="FeatureRecord" sawsdl:modelReference="http://edamontology.org/data_2044 http://edamontology.org/data_0867 http://edamontology.org/data_1381 http://edamontology.org/data_0855 http://edamontology.org/data_2201 http://edamontology.org/data_0924 http://edamontology.org/data_0857 http://edamontology.org/data_0916 http://edamontology.org/data_2084 http://edamontology.org/data_0896 http://edamontology.org/data_2081 http://edamontology.org/format_2352"> <xs:annotation> <xs:documentation>Features of a sequence/biopolymer (or a genome/metagenome). The reference sequence may me included explicitly, referred to remotely, or implicit</xs:documentation> </xs:annotation> <xs:sequence> <!--= Reference sequence =--> <xs:element name="referenceSequence"> <xs:annotation> <xs:documentation>The *reference* sequence that is annotated</xs:documentation> </xs:annotation> <xs:complexType> <xs:choice> <xs:element name="sequenceRecord" type="BiosequenceRecord"/> <xs:element name="sequenceReference" type="SequenceReference"/> <xs:element name="sequenceName" type="Name" sawsdl:modelReference="http://edamontology.org/data_2154"/> <xs:sequence> <xs:element name="species" type="Species"/> <xs:element name="genomeBuild" type="EntryReference" sawsdl:modelReference="http://edamontology.org/data_2340"/> <xs:element name="chromosome" type="ChromosomeName"/> <xs:element name="subsequencePosition" type="SequencePosition" minOccurs="0"> <xs:annotation> <xs:documentation>Coordinates of a sub-sequence within the chromosome</xs:documentation> </xs:annotation> </xs:element> <xs:element name="variantAccession" type="Accession" minOccurs="0" sawsdl:modelReference="http://edamontology.org/data_1063 http://edamontology.org/data_2091"/> <xs:element name="variantNote" type="Name" minOccurs="0" sawsdl:modelReference="http://edamontology.org/data_1063"/> </xs:sequence> </xs:choice> </xs:complexType> </xs:element> <xs:choice maxOccurs="unbounded"> <!--= Annotation by a feature (a track) =--> <xs:element name="annotation" sawsdl:modelReference="http://edamontology.org/data_3002 http://edamontology.org/format_2352"> <xs:annotation> <xs:documentation>Annotation by a particular feature or property. Corresponds to one sequence annotation track</xs:documentation> </xs:annotation> <!--"Sequence annotation track"(3002)--> <!--"BioXSD format"(2352)--> <xs:complexType> <xs:sequence> <!--= Feature type =--> <xs:element name="feature" type="FeatureType"/> <!--= Condensed references for feature occurrence(s) =--> <xs:element name="condensedReferences" minOccurs="0"> <xs:complexType> <xs:sequence> <xs:element name="method" type="Method" minOccurs="0"/> <xs:element name="ontology" type="OntologyReference" minOccurs="0"/> <xs:element name="database" type="DatabaseReference" minOccurs="0"/> <xs:element name="scoreType" type="ScoreType" minOccurs="0"/> </xs:sequence> </xs:complexType> </xs:element> <!--= Feature occurrence(s) (instance(s)) =--> <xs:element name="occurrence" type="FeatureOccurrenceData" minOccurs="0" maxOccurs="unbounded"/> </xs:sequence> </xs:complexType> </xs:element> <!--= Annotation by a block of inter-related features =--> <xs:element name="blockWithOccurrenceReferences"> <xs:complexType> <xs:sequence> <!--= Sources (globally declared) =--> <xs:element name="database" minOccurs="0" maxOccurs="unbounded" sawsdl:modelReference="http://edamontology.org/data_0856"> <xs:complexType> <xs:complexContent> <xs:extension base="DatabaseReference"> <xs:attribute name="localId" type="LocalId" use="required"/> </xs:extension> </xs:complexContent> </xs:complexType> </xs:element> <xs:element name="ontology" minOccurs="0" maxOccurs="unbounded"> <xs:complexType> <xs:complexContent> <xs:extension base="OntologyReference"> <xs:attribute name="localId" type="LocalId" use="required"/> </xs:extension> </xs:complexContent> </xs:complexType> </xs:element> <xs:element name="semanticConcept" minOccurs="0" maxOccurs="unbounded"> <xs:complexType> <xs:complexContent> <xs:extension base="SemanticConcept"> <xs:attribute name="localId" type="LocalId" use="required"/> </xs:extension> </xs:complexContent> </xs:complexType> </xs:element> <xs:element name="method" minOccurs="0" maxOccurs="unbounded" sawsdl:modelReference="http://edamontology.org/data_0856"> <xs:complexType> <xs:complexContent> <xs:extension base="Method"> <xs:attribute name="localId" type="LocalId" use="required"/> </xs:extension> </xs:complexContent> </xs:complexType> </xs:element> <xs:element name="scoreType" minOccurs="0" maxOccurs="unbounded"> <xs:complexType> <xs:complexContent> <xs:extension base="ScoreType"> <xs:attribute name="localId" type="LocalId" use="required"/> </xs:extension> </xs:complexContent> </xs:complexType> </xs:element> <!--= Annotation by a feature (in a block) =--> <xs:element name="annotation" maxOccurs="unbounded" sawsdl:modelReference="http://edamontology.org/data_3002 http://edamontology.org/format_2352"> <xs:annotation> <xs:documentation>Annotation by a particular feature or property. Corresponds to one sequence annotation track</xs:documentation> </xs:annotation> <!--"Sequence annotation track"(3002)--> <!--"BioXSD format"(2352)--> <xs:complexType> <xs:sequence> <!--= Feature type (in a block) =--> <xs:element name="feature" type="FeatureType"/> <!--= Condensed references for feature occurrence(s) =--> <xs:element name="condensedReferences" minOccurs="0"> <xs:complexType> <xs:sequence> <xs:choice> <xs:element name="method" type="Method" minOccurs="0"/> <xs:element name="methodIdRef" type="LocalId" minOccurs="0"/> </xs:choice> <xs:choice> <xs:element name="ontology" type="OntologyReference" minOccurs="0"/> <xs:element name="ontologyIdRef" type="LocalId" minOccurs="0"/> </xs:choice> <xs:choice> <xs:element name="database" type="DatabaseReference" minOccurs="0"/> <xs:element name="databaseIdRef" type="LocalId" minOccurs="0"/> </xs:choice> <xs:choice> <xs:element name="scoreType" type="ScoreType" minOccurs="0"/> <xs:element name="scoreTypeIdRef" type="LocalId" minOccurs="0"/> </xs:choice> </xs:sequence> </xs:complexType> </xs:element> <!--= Feature occurrence(s) (instance(s)) (in a block) =--> <xs:element name="occurrence" minOccurs="0" maxOccurs="unbounded"> <xs:complexType> <xs:complexContent> <xs:extension base="FeatureOccurrenceData"> <xs:sequence> <xs:element name="association" minOccurs="0" maxOccurs="unbounded"> <xs:complexType> <xs:sequence> <xs:element name="relationName" type="Name" minOccurs="0"/> <xs:element name="relationConcept" type="SemanticConcept" minOccurs="0" maxOccurs="unbounded"/> <xs:element name="relationConceptIdRef" type="LocalId" minOccurs="0" maxOccurs="unbounded"/> <xs:element name="associatedFeatureOccurrence" maxOccurs="unbounded"> <xs:complexType> <xs:sequence> <xs:element name="score" type="Score" minOccurs="0" maxOccurs="unbounded"/> </xs:sequence> <xs:attribute name="localIdRef" type="LocalId" use="required"/> </xs:complexType> </xs:element> </xs:sequence> </xs:complexType> </xs:element> </xs:sequence> <xs:attribute name="localId" type="LocalId"/> </xs:extension> </xs:complexContent> </xs:complexType> </xs:element> </xs:sequence> </xs:complexType> </xs:element> </xs:sequence> </xs:complexType> <xs:unique name="uniqueLocalId_occ"> <xs:selector xpath="annotation/occurrence"/> <xs:field xpath="@localId"/> </xs:unique> </xs:element> </xs:choice> </xs:sequence> <!--"Sequence data"(2044)--> <!--"Sequence alignment annotation"(0867)--> <!--"Sequence alignment (pairwise)"(1381)--> <!--"Sequence annotation"(0855)--> <!--"Sequence record full"(2201)--> <!--"Sequence trace"(0924)--> <!--"Sequence database hits"(0857)--> <!--"Gene annotation"(0916)--> <!--"Nucleic acid annotation"(2084)--> <!--"Protein annotation"(0896)--> <!--"Secondary structure"(2081)--> <!--"BioXSD format"(2352)--> </xs:complexType> |
| model reference | http://edamontology.org/data_2044 http://edamontology.org/data_0867 http://edamontology.org/data_1381 http://edamontology.org/data_0855 http://edamontology.org/data_2201 http://edamontology.org/data_0924 http://edamontology.org/data_0857 http://edamontology.org/data_0916 http://edamontology.org/data_2084 http://edamontology.org/data_0896 http://edamontology.org/data_2081 http://edamontology.org/format_2352 |
Evidence complexType
| diagram | 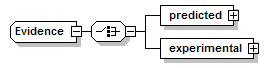 |
||
| children | predicted experimental | ||
| used by |
|
||
| source | <xs:complexType name="Evidence" sawsdl:modelReference="http://edamontology.org/data_2042 http://edamontology.org/format_2352"> <xs:choice> <xs:element name="predicted" type="PredictionResult"/> <xs:element name="experimental" type="ExperimentalEvidence"/> </xs:choice> </xs:complexType> |
||
| model reference | http://edamontology.org/data_2042 http://edamontology.org/format_2352 |
MethodResult complexType
| diagram |  |
| children | method score |
| used by | |
| source | <xs:complexType name="MethodResult"> <xs:sequence> <xs:element name="method"> <xs:complexType> <xs:complexContent> <xs:extension base="Method"> <xs:attribute name="localId" type="LocalId"/> </xs:extension> </xs:complexContent> </xs:complexType> </xs:element> <xs:element name="score" type="Score" minOccurs="0" maxOccurs="unbounded"/> </xs:sequence> </xs:complexType> |
Result complexType
| diagram | 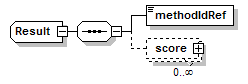 |
| children | methodIdRef score |
| used by | |
| source | <xs:complexType name="Result"> <xs:sequence> <xs:element name="methodIdRef" type="LocalId" sawsdl:modelReference="http://edamontology.org/data_0007 http://edamontology.org/data_2337 http://edamontology.org/data_0977 http://edamontology.org/format_2352"/> <xs:element name="score" type="Score" minOccurs="0" maxOccurs="unbounded"/> </xs:sequence> </xs:complexType> |
PredictionResult complexType
| diagram | 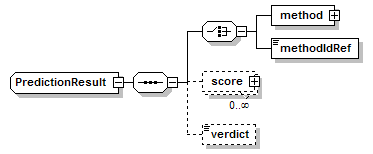 |
||
| children | method methodIdRef score verdict | ||
| used by |
|
||
| source | <xs:complexType name="PredictionResult"> <xs:sequence> <xs:choice> <xs:element name="method" type="Method" sawsdl:modelReference="http://edamontology.org/data_0856"/> <xs:element name="methodIdRef" type="LocalId" sawsdl:modelReference="http://edamontology.org/data_0007 http://edamontology.org/data_2337 http://edamontology.org/data_0977 http://edamontology.org/format_2352 http://edamontology.org/data_0856"/> </xs:choice> <xs:element name="score" type="Score" minOccurs="0" maxOccurs="unbounded"/> <xs:element name="verdict" type="Verdict" minOccurs="0"/> </xs:sequence> </xs:complexType> |
ExperimentalEvidence complexType
| diagram |  |
||
| children | dataReference citation verdict reliability | ||
| used by |
|
||
| source | <xs:complexType name="ExperimentalEvidence"> <xs:sequence> <xs:element name="dataReference" type="EntryReference" sawsdl:modelReference="http://edamontology.org/data_0856"/> <xs:element name="citation" type="EntryReference" minOccurs="0" maxOccurs="unbounded" sawsdl:modelReference="http://edamontology.org/data_0970 "/> <xs:element name="verdict" type="Verdict" minOccurs="0"/> <xs:element name="reliability" type="Reliability" minOccurs="0"/> </xs:sequence> </xs:complexType> |
ScoreType complexType
| diagram | 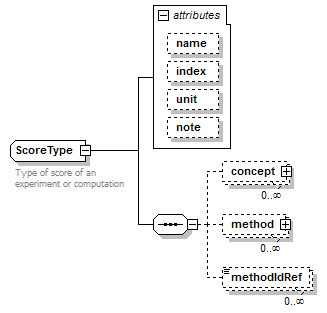 |
||||||||||||||||||||||||||||||
| children | concept method methodIdRef | ||||||||||||||||||||||||||||||
| used by | |||||||||||||||||||||||||||||||
| attributes |
|
||||||||||||||||||||||||||||||
| documentation | Type of score of an experiment or computation | ||||||||||||||||||||||||||||||
| source | <xs:complexType name="ScoreType" sawsdl:modelReference="http://edamontology.org/data_1772 http://edamontology.org/format_2352"> <xs:annotation> <xs:documentation>Type of score of an experiment or computation</xs:documentation> </xs:annotation> <xs:sequence> <xs:element name="concept" type="SemanticConcept" minOccurs="0" maxOccurs="unbounded"/> <xs:element name="method" type="Method" minOccurs="0" maxOccurs="unbounded" sawsdl:modelReference="http://edamontology.org/data_0856"/> <xs:element name="methodIdRef" type="LocalId" minOccurs="0" maxOccurs="unbounded" sawsdl:modelReference="http://edamontology.org/data_0007 http://edamontology.org/data_2337 http://edamontology.org/data_0977 http://edamontology.org/format_2352 http://edamontology.org/data_0856"/> </xs:sequence> <xs:attribute name="name" type="ScoreName"/> <xs:attribute name="index" type="Name"/> <xs:attribute name="unit" type="Name"/> <xs:attribute name="note" type="Text" sawsdl:modelReference="http://www.w3.org/2000/01/rdf-schema#comment"/> </xs:complexType> |
||||||||||||||||||||||||||||||
| model reference | http://edamontology.org/data_1772 http://edamontology.org/format_2352 |
Score complexType
| diagram | 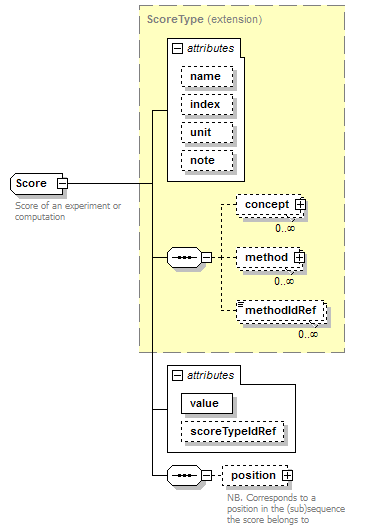 |
||||||||||||||||||||||||||||||||||||||||||
| type | extension of ScoreType | ||||||||||||||||||||||||||||||||||||||||||
| properties |
|
||||||||||||||||||||||||||||||||||||||||||
| children | concept method methodIdRef position | ||||||||||||||||||||||||||||||||||||||||||
| used by | |||||||||||||||||||||||||||||||||||||||||||
| attributes |
|
||||||||||||||||||||||||||||||||||||||||||
| documentation | Score of an experiment or computation | ||||||||||||||||||||||||||||||||||||||||||
| source | <xs:complexType name="Score" sawsdl:modelReference="http://edamontology.org/data_1772 http://edamontology.org/format_2352"> <xs:annotation> <xs:documentation>Score of an experiment or computation</xs:documentation> </xs:annotation> <xs:complexContent> <xs:extension base="ScoreType"> <xs:sequence> <xs:element name="position" type="SequencePosition" minOccurs="0"> <xs:annotation> <xs:documentation>NB. Corresponds to a position in the (sub)sequence the score belongs to</xs:documentation> </xs:annotation> </xs:element> </xs:sequence> <xs:attribute name="value" type="ScoreValue" use="required"/> <xs:attribute name="scoreTypeIdRef" type="LocalId"/> </xs:extension> </xs:complexContent> </xs:complexType> |
||||||||||||||||||||||||||||||||||||||||||
| model reference | http://edamontology.org/data_1772 http://edamontology.org/format_2352 |
GeneralSequencePoint complexType
| diagram | 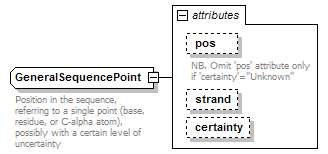 |
||||||||||||||||||||||||||
| used by |
|
||||||||||||||||||||||||||
| attributes |
|
||||||||||||||||||||||||||
| documentation | Position in the sequence, referring to a single point (base, residue, or C-alpha atom), possibly with a certain level of uncertainty | ||||||||||||||||||||||||||
| source | <xs:complexType name="GeneralSequencePoint" sawsdl:modelReference="http://edamontology.org/data_1016 http://edamontology.org/format_2352"> <xs:annotation> <xs:documentation>Position in the sequence, referring to a single point (base, residue, or C-alpha atom), possibly with a certain level of uncertainty</xs:documentation> </xs:annotation> <xs:attribute name="pos" type="PositiveInteger"> <xs:annotation> <xs:documentation>NB. Omit 'pos' attribute only if 'certainty'="Unknown"</xs:documentation> </xs:annotation> </xs:attribute> <xs:attribute name="strand" type="Strand"/> <xs:attribute name="certainty" type="Certainty" default="Certain"/> </xs:complexType> |
||||||||||||||||||||||||||
| model reference | http://edamontology.org/data_1016 http://edamontology.org/format_2352 |
SequencePoint complexType
| diagram | 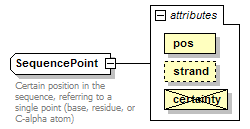 |
||||||||||||||||||||||||
| type | restriction of GeneralSequencePoint | ||||||||||||||||||||||||
| properties |
|
||||||||||||||||||||||||
| used by |
|
||||||||||||||||||||||||
| attributes |
|
||||||||||||||||||||||||
| documentation | Certain position in the sequence, referring to a single point (base, residue, or C-alpha atom) | ||||||||||||||||||||||||
| source | <xs:complexType name="SequencePoint"> <xs:annotation> <xs:documentation>Certain position in the sequence, referring to a single point (base, residue, or C-alpha atom)</xs:documentation> </xs:annotation> <xs:complexContent> <xs:restriction base="GeneralSequencePoint"> <xs:attribute name="pos" type="PositiveInteger" use="required"/> <xs:attribute name="certainty" type="Certainty" use="prohibited"/> </xs:restriction> </xs:complexContent> </xs:complexType> |
GeneralSequenceInsertionPoint complexType
| diagram | 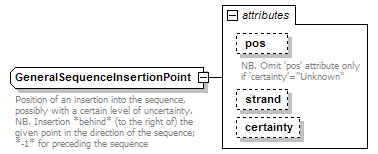 |
||||||||||||||||||||||||||
| used by |
|
||||||||||||||||||||||||||
| attributes |
|
||||||||||||||||||||||||||
| documentation | Position of an insertion into the sequence, possibly with a certain level of uncertainty. NB. Insertion *behind* (to the right of) the given point in the direction of the sequence; *-1* for preceding the sequence |
||||||||||||||||||||||||||
| source | <xs:complexType name="GeneralSequenceInsertionPoint" sawsdl:modelReference="http://edamontology.org/data_1016"> <xs:annotation> <xs:documentation>Position of an insertion into the sequence, possibly with a certain level of uncertainty. NB. Insertion *behind* (to the right of) the given point in the direction of the sequence; *-1* for preceding the sequence</xs:documentation> </xs:annotation> <xs:attribute name="pos" type="InsertionInteger"> <xs:annotation> <xs:documentation>NB. Omit 'pos' attribute only if 'certainty'="Unknown"</xs:documentation> </xs:annotation> </xs:attribute> <xs:attribute name="strand" type="Strand"/> <xs:attribute name="certainty" type="Certainty" default="Certain"/> </xs:complexType> |
||||||||||||||||||||||||||
| model reference | http://edamontology.org/data_1016 |
SequenceInsertionPoint complexType
| diagram | 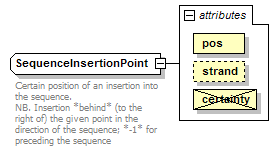 |
||||||||||||||||||||||||
| type | restriction of GeneralSequenceInsertionPoint | ||||||||||||||||||||||||
| properties |
|
||||||||||||||||||||||||
| used by |
|
||||||||||||||||||||||||
| attributes |
|
||||||||||||||||||||||||
| documentation | Certain position of an insertion into the sequence. NB. Insertion *behind* (to the right of) the given point in the direction of the sequence; *-1* for preceding the sequence |
||||||||||||||||||||||||
| source | <xs:complexType name="SequenceInsertionPoint"> <xs:annotation> <xs:documentation>Certain position of an insertion into the sequence. NB. Insertion *behind* (to the right of) the given point in the direction of the sequence; *-1* for preceding the sequence</xs:documentation> </xs:annotation> <xs:complexContent> <xs:restriction base="GeneralSequenceInsertionPoint"> <xs:attribute name="pos" type="InsertionInteger" use="required"/> <xs:attribute name="certainty" type="Certainty" use="prohibited"/> </xs:restriction> </xs:complexContent> </xs:complexType> |
GeneralOutsideSequencePoint complexType
| diagram | 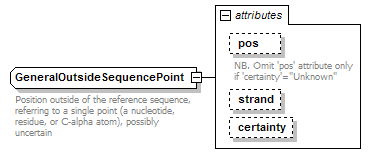 |
||||||||||||||||||||||||||
| used by |
|
||||||||||||||||||||||||||
| attributes |
|
||||||||||||||||||||||||||
| documentation | Position outside of the reference sequence, referring to a single point (a nucleotide, residue, or C-alpha atom), possibly uncertain | ||||||||||||||||||||||||||
| source | <xs:complexType name="GeneralOutsideSequencePoint" sawsdl:modelReference="http://edamontology.org/data_1016"> <xs:annotation> <xs:documentation>Position outside of the reference sequence, referring to a single point (a nucleotide, residue, or C-alpha atom), possibly uncertain</xs:documentation> </xs:annotation> <xs:attribute name="pos" type="NonzeroInteger"> <xs:annotation> <xs:documentation>NB. Omit 'pos' attribute only if 'certainty'="Unknown"</xs:documentation> </xs:annotation> </xs:attribute> <xs:attribute name="strand" type="Strand"/> <xs:attribute name="certainty" type="Certainty" default="Certain"/> </xs:complexType> |
||||||||||||||||||||||||||
| model reference | http://edamontology.org/data_1016 |
OutsideSequencePoint complexType
| diagram | 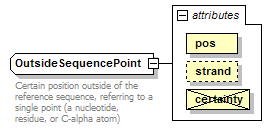 |
||||||||||||||||||||||||
| type | restriction of GeneralOutsideSequencePoint | ||||||||||||||||||||||||
| properties |
|
||||||||||||||||||||||||
| used by |
|
||||||||||||||||||||||||
| attributes |
|
||||||||||||||||||||||||
| documentation | Certain position outside of the reference sequence, referring to a single point (a nucleotide, residue, or C-alpha atom) | ||||||||||||||||||||||||
| source | <xs:complexType name="OutsideSequencePoint"> <xs:annotation> <xs:documentation>Certain position outside of the reference sequence, referring to a single point (a nucleotide, residue, or C-alpha atom)</xs:documentation> </xs:annotation> <xs:complexContent> <xs:restriction base="GeneralOutsideSequencePoint"> <xs:attribute name="pos" type="NonzeroInteger" use="required"/> <xs:attribute name="certainty" type="Certainty" use="prohibited"/> </xs:restriction> </xs:complexContent> </xs:complexType> |
GeneralSequenceSegment complexType
| diagram |  |
||||||||||||||||||||||||||||||||||||||||
| used by |
|
||||||||||||||||||||||||||||||||||||||||
| attributes |
|
||||||||||||||||||||||||||||||||||||||||
| documentation | Position in the sequence referring to a continuous segment of the sequence, possibly uncertain | ||||||||||||||||||||||||||||||||||||||||
| source | <xs:complexType name="GeneralSequenceSegment" sawsdl:modelReference="http://edamontology.org/data_1017"> <xs:annotation> <xs:documentation>Position in the sequence referring to a continuous segment of the sequence, possibly uncertain</xs:documentation> </xs:annotation> <xs:attribute name="min" type="PositiveInteger"> <xs:annotation> <xs:documentation>NB. Keep 'min' < 'max', use 'strand' if necessary. Omit 'min' attribute only if 'minCertainty'="Unknown"</xs:documentation> </xs:annotation> </xs:attribute> <xs:attribute name="max" type="PositiveInteger"> <xs:annotation> <xs:documentation>NB. Keep 'max' > 'min', use 'strand' if necessary. Omit 'max' attribute only if 'maxCertainty'="Unknown"</xs:documentation> </xs:annotation> </xs:attribute> <xs:attribute name="strand" type="Strand"/> <xs:attribute name="minCertainty" type="Certainty" default="Certain"/> <xs:attribute name="maxCertainty" type="Certainty" default="Certain"/> <!--"Sequence range"(1017)--> </xs:complexType> |
||||||||||||||||||||||||||||||||||||||||
| model reference | http://edamontology.org/data_1017 |
SequenceSegment complexType
| diagram | 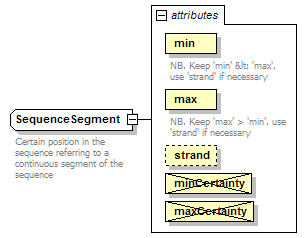 |
||||||||||||||||||||||||||||||||||||||||
| type | restriction of GeneralSequenceSegment | ||||||||||||||||||||||||||||||||||||||||
| properties |
|
||||||||||||||||||||||||||||||||||||||||
| used by |
|
||||||||||||||||||||||||||||||||||||||||
| attributes |
|
||||||||||||||||||||||||||||||||||||||||
| documentation | Certain position in the sequence referring to a continuous segment of the sequence | ||||||||||||||||||||||||||||||||||||||||
| source | <xs:complexType name="SequenceSegment"> <xs:annotation> <xs:documentation>Certain position in the sequence referring to a continuous segment of the sequence</xs:documentation> </xs:annotation> <xs:complexContent> <xs:restriction base="GeneralSequenceSegment"> <xs:attribute name="min" type="PositiveInteger" use="required"> <xs:annotation> <xs:documentation>NB. Keep 'min' < 'max', use 'strand' if necessary</xs:documentation> </xs:annotation> </xs:attribute> <xs:attribute name="max" type="PositiveInteger" use="required"> <xs:annotation> <xs:documentation>NB. Keep 'max' > 'min', use 'strand' if necessary</xs:documentation> </xs:annotation> </xs:attribute> <xs:attribute name="minCertainty" type="Certainty" use="prohibited"/> <xs:attribute name="maxCertainty" type="Certainty" use="prohibited"/> </xs:restriction> </xs:complexContent> </xs:complexType> |
GeneralOutsideSequenceSegment complexType
| diagram |  |
||||||||||||||||||||||||||||||||||||||||
| used by |
|
||||||||||||||||||||||||||||||||||||||||
| attributes |
|
||||||||||||||||||||||||||||||||||||||||
| documentation | Position outside of the reference sequence, referring to a continuous segment, possibly uncertain | ||||||||||||||||||||||||||||||||||||||||
| source | <xs:complexType name="GeneralOutsideSequenceSegment" sawsdl:modelReference="http://edamontology.org/data_1017"> <xs:annotation> <xs:documentation>Position outside of the reference sequence, referring to a continuous segment, possibly uncertain</xs:documentation> </xs:annotation> <xs:attribute name="min" type="NonzeroInteger"> <xs:annotation> <xs:documentation>NB. Keep 'min' < 'max', use 'strand' if necessary. Omit 'min' attribute only if 'minCertainty'="Unknown"</xs:documentation> </xs:annotation> </xs:attribute> <xs:attribute name="max" type="NonzeroInteger"> <xs:annotation> <xs:documentation>NB. Keep 'max' > 'min', use 'strand' if necessary. Omit 'max' attribute only if 'maxCertainty'="Unknown"</xs:documentation> </xs:annotation> </xs:attribute> <xs:attribute name="strand" type="Strand"/> <xs:attribute name="minCertainty" type="Certainty" default="Certain"/> <xs:attribute name="maxCertainty" type="Certainty" default="Certain"/> </xs:complexType> |
||||||||||||||||||||||||||||||||||||||||
| model reference | http://edamontology.org/data_1017 |
OutsideSequenceSegment complexType
| diagram | 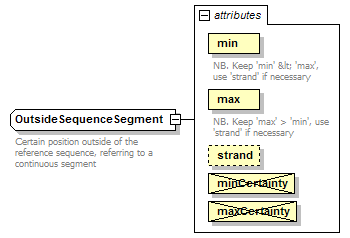 |
||||||||||||||||||||||||||||||||||||||||
| type | restriction of GeneralOutsideSequenceSegment | ||||||||||||||||||||||||||||||||||||||||
| properties |
|
||||||||||||||||||||||||||||||||||||||||
| used by |
|
||||||||||||||||||||||||||||||||||||||||
| attributes |
|
||||||||||||||||||||||||||||||||||||||||
| documentation | Certain position outside of the reference sequence, referring to a continuous segment | ||||||||||||||||||||||||||||||||||||||||
| source | <xs:complexType name="OutsideSequenceSegment"> <xs:annotation> <xs:documentation>Certain position outside of the reference sequence, referring to a continuous segment</xs:documentation> </xs:annotation> <xs:complexContent> <xs:restriction base="GeneralOutsideSequenceSegment"> <xs:attribute name="min" type="NonzeroInteger" use="required"> <xs:annotation> <xs:documentation>NB. Keep 'min' < 'max', use 'strand' if necessary</xs:documentation> </xs:annotation> </xs:attribute> <xs:attribute name="max" type="NonzeroInteger" use="required"> <xs:annotation> <xs:documentation>NB. Keep 'max' > 'min', use 'strand' if necessary</xs:documentation> </xs:annotation> </xs:attribute> <xs:attribute name="minCertainty" type="Certainty" use="prohibited"/> <xs:attribute name="maxCertainty" type="Certainty" use="prohibited"/> </xs:restriction> </xs:complexContent> </xs:complexType> |
GeneralSequencePartition complexType
| diagram | 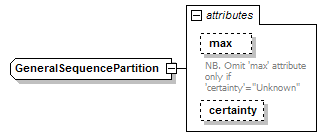 |
||||||||||||||||||||
| used by |
|
||||||||||||||||||||
| attributes |
|
||||||||||||||||||||
| source | <xs:complexType name="GeneralSequencePartition"> <xs:attribute name="max" type="PositiveInteger"> <xs:annotation> <xs:documentation>NB. Omit 'max' attribute only if 'certainty'="Unknown"</xs:documentation> </xs:annotation> </xs:attribute> <xs:attribute name="certainty" type="Certainty" default="Certain"/> </xs:complexType> |
SequencePartition complexType
| diagram | 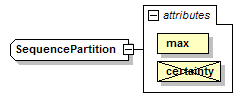 |
||||||||||||||||||
| type | restriction of GeneralSequencePartition | ||||||||||||||||||
| properties |
|
||||||||||||||||||
| used by |
|
||||||||||||||||||
| attributes |
|
||||||||||||||||||
| source | <xs:complexType name="SequencePartition"> <xs:complexContent> <xs:restriction base="GeneralSequencePartition"> <xs:attribute name="max" type="PositiveInteger" use="required"/> <xs:attribute name="certainty" type="Certainty" use="prohibited"/> </xs:restriction> </xs:complexContent> </xs:complexType> |
GeneralSequencePosition complexType
| diagram | 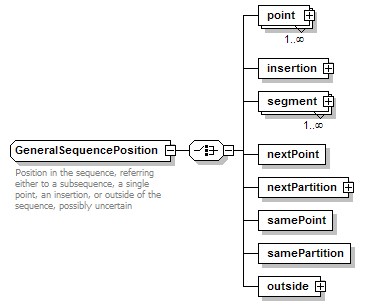 |
||||
| children | point insertion segment nextPoint nextPartition samePoint samePartition outside | ||||
| used by |
|
||||
| documentation | Position in the sequence, referring either to a subsequence, a single point, an insertion, or outside of the sequence, possibly uncertain | ||||
| source | <xs:complexType name="GeneralSequencePosition" sawsdl:modelReference="http://edamontology.org/data_1014"> <xs:annotation> <xs:documentation>Position in the sequence, referring either to a subsequence, a single point, an insertion, or outside of the sequence, possibly uncertain</xs:documentation> </xs:annotation> <xs:choice> <xs:element name="point" type="GeneralSequencePoint" maxOccurs="unbounded"/> <xs:element name="insertion" type="GeneralSequenceInsertionPoint"/> <xs:element name="segment" type="GeneralSequenceSegment" maxOccurs="unbounded"/> <xs:element name="nextPoint" type="Flag"/> <xs:element name="nextPartition" type="GeneralSequencePartition"/> <xs:element name="samePoint" type="Flag"/> <xs:element name="samePartition" type="Flag"/> <xs:element name="outside" type="GeneralOutsideSequencePosition"/> </xs:choice> <!--"Sequence position specification"(1014)--> </xs:complexType> |
||||
| model reference | http://edamontology.org/data_1014 |
SequencePosition complexType
| diagram | 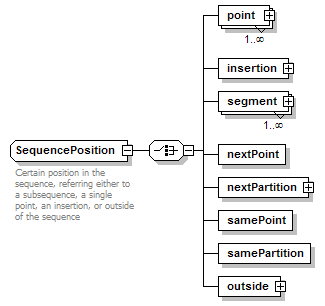 |
||
| type | restriction of GeneralSequencePosition | ||
| properties |
|
||
| children | point insertion segment nextPoint nextPartition samePoint samePartition outside | ||
| used by | |||
| documentation | Certain position in the sequence, referring either to a subsequence, a single point, an insertion, or outside of the sequence | ||
| source | <xs:complexType name="SequencePosition"> <xs:annotation> <xs:documentation>Certain position in the sequence, referring either to a subsequence, a single point, an insertion, or outside of the sequence</xs:documentation> </xs:annotation> <xs:complexContent> <xs:restriction base="GeneralSequencePosition"> <xs:choice> <xs:element name="point" type="SequencePoint" maxOccurs="unbounded"/> <xs:element name="insertion" type="SequenceInsertionPoint"/> <xs:element name="segment" type="SequenceSegment" maxOccurs="unbounded"/> <xs:element name="nextPoint" type="Flag"/> <xs:element name="nextPartition" type="SequencePartition"/> <xs:element name="samePoint" type="Flag"/> <xs:element name="samePartition" type="Flag"/> <xs:element name="outside" type="OutsideSequencePosition"/> </xs:choice> </xs:restriction> </xs:complexContent> </xs:complexType> |
GeneralOutsideSequencePosition complexType
| diagram | 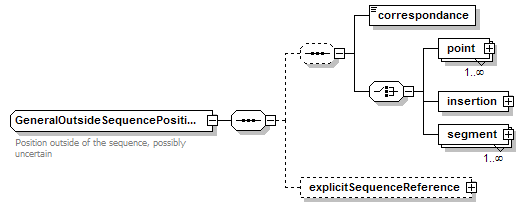 |
||||
| children | correspondance point insertion segment explicitSequenceReference | ||||
| used by |
|
||||
| documentation | Position outside of the sequence, possibly uncertain | ||||
| source | <xs:complexType name="GeneralOutsideSequencePosition" sawsdl:modelReference="http://edamontology.org/data_1014"> <xs:annotation> <xs:documentation>Position outside of the sequence, possibly uncertain</xs:documentation> </xs:annotation> <xs:sequence> <xs:sequence minOccurs="0"> <xs:element name="correspondance" type="OutsidePositionCorrespondance"/> <xs:choice> <xs:element name="point" type="GeneralOutsideSequencePoint" maxOccurs="unbounded"/> <xs:element name="insertion" type="GeneralOutsideSequencePoint"/> <xs:element name="segment" type="GeneralOutsideSequenceSegment" maxOccurs="unbounded"/> </xs:choice> </xs:sequence> <xs:element name="explicitSequenceReference" type="SequenceReference" minOccurs="0"/> </xs:sequence> </xs:complexType> |
||||
| model reference | http://edamontology.org/data_1014 |
OutsideSequencePosition complexType
| diagram | 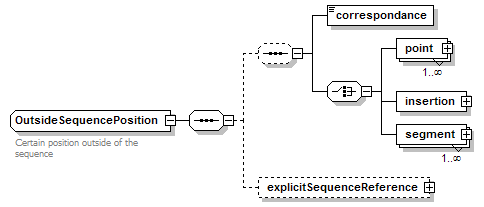 |
||
| type | restriction of GeneralOutsideSequencePosition | ||
| properties |
|
||
| children | correspondance point insertion segment explicitSequenceReference | ||
| used by |
|
||
| documentation | Certain position outside of the sequence | ||
| source | <xs:complexType name="OutsideSequencePosition"> <xs:annotation> <xs:documentation>Certain position outside of the sequence</xs:documentation> </xs:annotation> <xs:complexContent> <xs:restriction base="GeneralOutsideSequencePosition"> <xs:sequence> <xs:sequence minOccurs="0"> <xs:element name="correspondance" type="OutsidePositionCorrespondance"/> <xs:choice> <xs:element name="point" type="OutsideSequencePoint" maxOccurs="unbounded"/> <xs:element name="insertion" type="OutsideSequencePoint"/> <xs:element name="segment" type="OutsideSequenceSegment" maxOccurs="unbounded"/> </xs:choice> </xs:sequence> <xs:element name="explicitSequenceReference" type="SequenceReference" minOccurs="0"/> </xs:sequence> </xs:restriction> </xs:complexContent> </xs:complexType> |
GeneticCode complexType
| diagram |  |
||
| children | reference codon | ||
| used by |
|
||
| documentation | Particular genetic code (codon encoding) | ||
| source | <xs:complexType name="GeneticCode" sawsdl:modelReference="http://edamontology.org/data_1598 http://edamontology.org/format_2352"> <xs:annotation> <xs:documentation>Particular genetic code (codon encoding)</xs:documentation> </xs:annotation> <xs:sequence> <xs:element name="reference" type="EntryReference" minOccurs="0" sawsdl:modelReference="http://edamontology.org/data_2127"/> <xs:element name="codon" minOccurs="0" maxOccurs="unbounded" sawsdl:modelReference="http://edamontology.org/data_2116 http://edamontology.org/format_2352"> <xs:complexType> <xs:attribute name="code" use="required"> <xs:annotation> <xs:documentation>Codon code consisting of 3 bases (possibly ambiguous, "degenerate")</xs:documentation> </xs:annotation> <xs:simpleType> <xs:restriction base="GeneralNucleotideSequence"> <xs:length value="3"/> </xs:restriction> </xs:simpleType> </xs:attribute> <xs:attribute name="start" type="xs:boolean" default="false"/> <xs:attribute name="end" type="xs:boolean" default="false"/> <xs:attribute name="aminoacid" sawsdl:modelReference="http://edamontology.org/data_0994"> <xs:annotation> <xs:documentation>One amino-acid (possibly ambiguous). NB. If the same codon codes for multiple amino-acids, use multiple 'codon' elements and fill in the 'note' attribute</xs:documentation> </xs:annotation> <xs:simpleType> <xs:restriction base="GeneralAminoacidSequence"> <xs:length value="1"/> </xs:restriction> </xs:simpleType> </xs:attribute> <xs:attribute name="veryRare" type="xs:boolean" default="false"/> <xs:attribute name="note" type="Text" sawsdl:modelReference="http://www.w3.org/2000/01/rdf-schema#comment"/> </xs:complexType> </xs:element> </xs:sequence> </xs:complexType> |
||
| model reference | http://edamontology.org/data_1598 http://edamontology.org/format_2352 |
TranslationData complexType
| diagram | 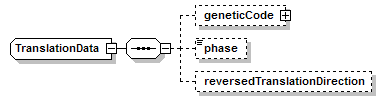 |
||||
| children | geneticCode phase reversedTranslationDirection | ||||
| used by |
|
||||
| source | <xs:complexType name="TranslationData" sawsdl:modelReference="http://edamontology.org/data_1317 http://edamontology.org/format_2352"> <xs:sequence> <xs:element name="geneticCode" type="GeneticCode" minOccurs="0"/> <xs:element name="phase" type="Phase" minOccurs="0"/> <xs:element name="reversedTranslationDirection" type="Flag" minOccurs="0"/> </xs:sequence> </xs:complexType> |
||||
| model reference | http://edamontology.org/data_1317 http://edamontology.org/format_2352 |
AminoacidTranslationData complexType
| diagram | |||
| type | restriction of TranslationData | ||
| properties |
|
||
| children | geneticCode | ||
| used by |
|
||
| source | <xs:complexType name="AminoacidTranslationData"> <xs:complexContent> <xs:restriction base="TranslationData"> <xs:sequence> <xs:element name="geneticCode" type="GeneticCode" minOccurs="0"/> </xs:sequence> </xs:restriction> </xs:complexContent> </xs:complexType> |
DatabaseReference complexType
| diagram |  |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| used by | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| attributes |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| documentation | Reference to a database | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| source | <xs:complexType name="DatabaseReference" sawsdl:modelReference="http://edamontology.org/format_2352"> <xs:annotation> <xs:documentation>Reference to a database</xs:documentation> </xs:annotation> <xs:attribute name="dbName" type="DatabaseName"/> <xs:attribute name="dbUri" type="Uri" sawsdl:modelReference="http://purl.org/dc/elements/1.1/identifier http://edamontology.org/data_1048"> <xs:annotation> <xs:documentation>General URI of the database</xs:documentation> </xs:annotation> <!--"Database identifier"(1048)--> <!--"Identifier"(0842)--> </xs:attribute> <xs:attribute name="dbVersion" type="Name" sawsdl:modelReference="http://purl.org/dc/elements/1.1/version http://edamontology.org/data_1670"/> <xs:attribute name="localCopyOfDb" type="xs:boolean" default="false"/> <xs:attributeGroup ref="WebserviceReference"/> <!--"Database identifier"(1048)--> <!--"Identifier"(0842)--> <!--"BioXSD"(2352)--> <!--"Database version information"(1670)--> </xs:complexType> |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| model reference | http://edamontology.org/format_2352 |
EntryReference complexType
| diagram | 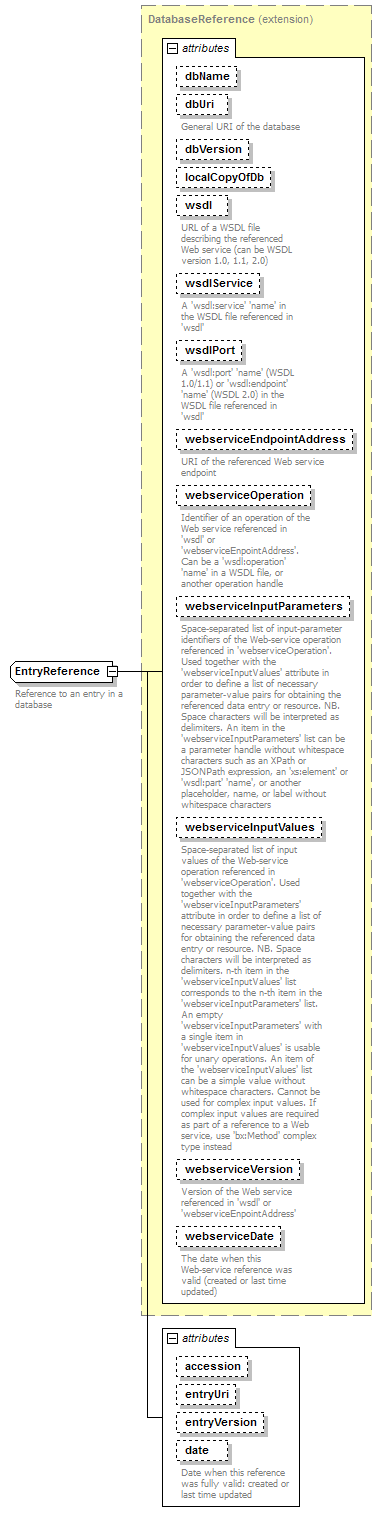 |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| type | extension of DatabaseReference | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| properties |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| used by | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| attributes |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| documentation | Reference to an entry in a database | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| source | <xs:complexType name="EntryReference"> <xs:annotation> <xs:documentation>Reference to an entry in a database</xs:documentation> </xs:annotation> <xs:complexContent> <xs:extension base="DatabaseReference"> <xs:attribute name="accession" type="Accession"/> <xs:attribute name="entryUri" type="Uri" sawsdl:modelReference="http://purl.org/dc/elements/1.1/identifier http://edamontology.org/data_2091"/> <xs:attribute name="entryVersion" type="Name" sawsdl:modelReference="http://edamontology.org/data_2090 http://purl.org/dc/elements/1.1/version"/> <xs:attribute name="date" type="xs:date" sawsdl:modelReference="http://edamontology.org/data_2156 http://edamontology.org/data_2337"> <xs:annotation> <xs:documentation>Date when this reference was fully valid: created or last time updated</xs:documentation> </xs:annotation> </xs:attribute> </xs:extension> <!--"Accession"(2091)--> <!--"Identifier"(0842)--> <!--"Database entry version information"(2090)--> </xs:complexContent> <!--"Identifier"(0842)--> </xs:complexType> |
OntologyReference complexType
| diagram | 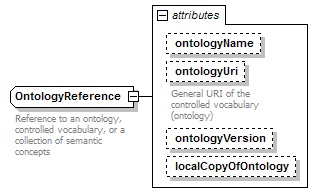 |
||||||||||||||||||||||||||||||||
| used by | |||||||||||||||||||||||||||||||||
| attributes |
|
||||||||||||||||||||||||||||||||
| documentation | Reference to an ontology, controlled vocabulary, or a collection of semantic concepts | ||||||||||||||||||||||||||||||||
| source | <xs:complexType name="OntologyReference" sawsdl:modelReference="http://edamontology.org/data_2338 http://edamontology.org/data_2352"> <xs:annotation> <xs:documentation>Reference to an ontology, controlled vocabulary, or a collection of semantic concepts</xs:documentation> </xs:annotation> <xs:attribute name="ontologyName" type="OntologyName" sawsdl:modelReference="http://edamontology.org/data_1051"/> <xs:attribute name="ontologyUri" type="Uri" sawsdl:modelReference="http://edamontology.org/data_2338"> <xs:annotation> <xs:documentation>General URI of the controlled vocabulary (ontology)</xs:documentation> </xs:annotation> </xs:attribute> <xs:attribute name="ontologyVersion" type="Name" sawsdl:modelReference="http://purl.org/dc/elements/1.1/version http://edamontology.org/data_0953"/> <xs:attribute name="localCopyOfOntology" type="xs:boolean" default="false"/> <!--"Version information"(0953)--> </xs:complexType> |
||||||||||||||||||||||||||||||||
| model reference | http://edamontology.org/data_2338 http://edamontology.org/data_2352 |
SemanticConcept complexType
| diagram | 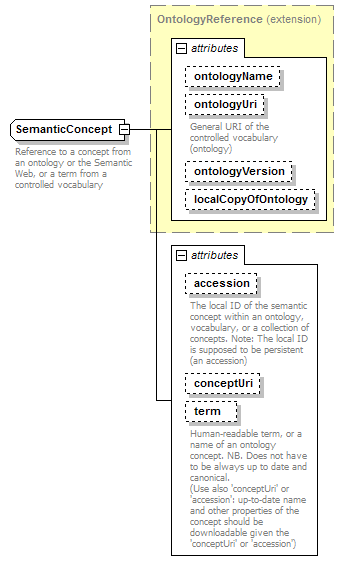 |
||||||||||||||||||||||||||||||||||||||||||||||||||||||
| type | extension of OntologyReference | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| properties |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||
| used by | |||||||||||||||||||||||||||||||||||||||||||||||||||||||
| attributes |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||
| documentation | Reference to a concept from an ontology or the Semantic Web, or a term from a controlled vocabulary | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| source | <xs:complexType name="SemanticConcept" sawsdl:modelReference="http://edamontology.org/data_0966 http://edamontology.org/data_1087 http://edamontology.org/data_2352"> <xs:annotation> <xs:documentation>Reference to a concept from an ontology or the Semantic Web, or a term from a controlled vocabulary</xs:documentation> </xs:annotation> <xs:complexContent> <xs:extension base="OntologyReference"> <xs:attribute name="accession" type="Accession" sawsdl:modelReference="http://edamontology.org/data_1087"> <xs:annotation> <xs:documentation>The local ID of the semantic concept within an ontology, vocabulary, or a collection of concepts. Note: The local ID is supposed to be persistent (an accession)</xs:documentation> </xs:annotation> </xs:attribute> <xs:attribute name="conceptUri" type="Uri" sawsdl:modelReference="http://edamontology.org/data_1087"/> <xs:attribute name="term" type="Name" sawsdl:modelReference="http://edamontology.org/data_2339"> <xs:annotation> <xs:documentation>Human-readable term, or a name of an ontology concept. NB. Does not have to be always up to date and canonical. (Use also 'conceptUri' or 'accession': up-to-date name and other properties of the concept should be downloadable given the 'conceptUri' or 'accession')</xs:documentation> </xs:annotation> </xs:attribute> </xs:extension> </xs:complexContent> </xs:complexType> |
||||||||||||||||||||||||||||||||||||||||||||||||||||||
| model reference | http://edamontology.org/data_0966 http://edamontology.org/data_1087 http://edamontology.org/data_2352 |
Species complexType
| diagram |  |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| type | extension of EntryReference | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| properties |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| used by | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| attributes |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| documentation | Reference to a species (can be used also for a phenotype, cell line, tissue, sample, geo location, ...) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| source | <xs:complexType name="Species"> <xs:annotation> <xs:documentation>Reference to a species (can be used also for a phenotype, cell line, tissue, sample, geo location, ...)</xs:documentation> </xs:annotation> <xs:complexContent> <xs:extension base="EntryReference"> <xs:attribute name="speciesName" type="Name" sawsdl:modelReference="http://www.w3.org/2000/01/rdf-schema#label http://edamontology.org/data_1045"> <xs:annotation> <xs:documentation>Custom human-readable name of the species. NB. Does not have to be always up to date and canonical. Use also entryUri or accession if possible</xs:documentation> </xs:annotation> <!--"Species name"(1045)--> </xs:attribute> <xs:attribute name="speciesNote" type="Text" sawsdl:modelReference="http://www.w3.org/2000/01/rdf-schema#comment"/> </xs:extension> </xs:complexContent> <!--"Organism identifier"(1869)--> </xs:complexType> |
SequenceReference complexType
| diagram |  |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| type | extension of EntryReference | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| properties |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| children | subsequencePosition supersequence supersequenceName | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| used by | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| attributes |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| documentation | Formal reference to a sequence in a database or an explicit super-sequence | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| source | <xs:complexType name="SequenceReference"> <xs:annotation> <xs:documentation>Formal reference to a sequence in a database or an explicit super-sequence</xs:documentation> </xs:annotation> <xs:complexContent> <xs:extension base="EntryReference"> <xs:sequence> <xs:element name="subsequencePosition" type="SequencePosition" minOccurs="0"> <xs:annotation> <xs:documentation>Coordinates of the sub-sequence within the referenced sequence</xs:documentation> </xs:annotation> </xs:element> <xs:element name="supersequence" type="Biosequence" minOccurs="0"> <xs:annotation> <xs:documentation>Explicit super-sequence, in case it is not desired or not possible to point to a database entry</xs:documentation> </xs:annotation> </xs:element> <xs:element name="supersequenceName" type="Name" minOccurs="0" sawsdl:modelReference="http://edamontology.org/data_2154"> <xs:annotation> <xs:documentation>Custom name of the super-sequence, in case it is not desired or not possible to point to a database entry</xs:documentation> </xs:annotation> </xs:element> </xs:sequence> <xs:attribute name="sequenceVersion" type="Name" sawsdl:modelReference="http://edamontology.org/data_1771 http://purl.org/dc/elements/1.1/version"/> <xs:attribute name="variantAccession" type="Accession" sawsdl:modelReference="http://purl.org/dc/elements/1.1/identifier http://edamontology.org/data_1063"/> <xs:attribute name="variantNote" type="Name" sawsdl:modelReference="http://www.w3.org/2000/01/rdf-schema#comment http://edamontology.org/data_1063"/> </xs:extension> <!--"Sequence version information"(1771)--> <!--"Sequence identifier"(1063)--> </xs:complexContent> <!--"Sequence identifier"(1063)--> </xs:complexType> |
Method complexType
| diagram |  |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| children | categoryConcept citation inputParameter outputParameter | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| used by | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| attributes |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| source | <xs:complexType name="Method" sawsdl:modelReference="http://edamontology.org/data_0007 http://edamontology.org/data_2337 http://edamontology.org/data_0977 http://edamontology.org/format_2352"> <xs:sequence> <xs:element name="categoryConcept" type="SemanticConcept" minOccurs="0" maxOccurs="unbounded" sawsdl:modelReference="http://edamontology.org/data_0958 http://edamontology.org/operation_0004"/> <xs:element name="citation" type="EntryReference" minOccurs="0" maxOccurs="unbounded" sawsdl:modelReference="http://edamontology.org/data_0958 http://edamontology.org/data_0970 "/> <xs:element name="inputParameter" type="Score" minOccurs="0" maxOccurs="unbounded" sawsdl:modelReference="http://edamontology.org/data_2527"/> <!--"Parameter"(2527)--> <xs:element name="outputParameter" type="Score" minOccurs="0" maxOccurs="unbounded" sawsdl:modelReference="http://edamontology.org/data_2527"/> <!--"Parameter"(2527)--> </xs:sequence> <xs:attribute name="name" type="Name" use="required" sawsdl:modelReference="http://edamontology.org/data_1190"/> <xs:attribute name="uri" type="Uri" sawsdl:modelReference="http://edamontology.org/data_0977"/> <xs:attribute name="version" type="Name" sawsdl:modelReference="http://purl.org/dc/elements/1.1/version http://edamontology.org/data_1671"> <!--"Tool version information"(1671)--> </xs:attribute> <xs:attribute name="date" type="xs:date" sawsdl:modelReference="http://edamontology.org/data_2156 http://edamontology.org/data_2337"/> <xs:attributeGroup ref="WebserviceReference"/> </xs:complexType> |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| model reference | http://edamontology.org/data_0007 http://edamontology.org/data_2337 http://edamontology.org/data_0977 http://edamontology.org/format_2352 |
LocalId simpleType
Accession simpleType
| type | restriction of xs:string | |||||||||
| used by |
|
|||||||||
| facets |
|
|||||||||
| documentation | Generalisation of bioinformatics accession numbers (stable primary keys/stable unique identifiers), or bioinformatics primary keys/unique identifiers (where no stable accessions) | |||||||||
| source | <xs:simpleType name="Accession" sawsdl:modelReference="http://purl.org/dc/elements/1.1/identifier http://edamontology.org/data_0842 http://edamontology.org/data_2091"> <xs:annotation> <xs:documentation>Generalisation of bioinformatics accession numbers (stable primary keys/stable unique identifiers), or bioinformatics primary keys/unique identifiers (where no stable accessions)</xs:documentation> </xs:annotation> <xs:restriction base="xs:string"> <xs:maxLength value="1000"/> <xs:pattern value="[A-Za-z0-9\-._~]+"/> </xs:restriction> <!--"Identifier"(0842)--> <!--"Accession"(2091)--> </xs:simpleType> |
EmblAccession simpleType
| type | Accession | |||||||||
| facets |
|
|||||||||
| source | <xs:simpleType name="EmblAccession" sawsdl:modelReference="http://edamontology.org/data_1107"> <xs:restriction base="Accession"/> </xs:simpleType> |
ExtendedUniprotAccession simpleType
| type | restriction of Accession | ||||||||||||||||||||||||
| used by |
|
||||||||||||||||||||||||
| facets |
|
||||||||||||||||||||||||
| documentation | UniProt accession number, optionally with version after a dot (.) or the variant (isoform) suffix after a hyphen (-) | ||||||||||||||||||||||||
| source | <xs:simpleType name="ExtendedUniprotAccession"> <xs:annotation> <xs:documentation>UniProt accession number, optionally with version after a dot (.) or the variant (isoform) suffix after a hyphen (-)</xs:documentation> </xs:annotation> <xs:restriction base="Accession"> <xs:pattern value="[OPQ][0-9][A-Z0-9][A-Z0-9][A-Z0-9][0-9]"/> <xs:pattern value="[A-NR-Z][0-9][A-Z][A-Z0-9][A-Z0-9][0-9]"/> <xs:pattern value="[A-NR-Z][0-9][A-Z][A-Z0-9][A-Z0-9][0-9][A-Z][A-Z0-9][A-Z0-9][0-9]"/> <xs:pattern value="[OPQ][0-9][A-Z0-9][A-Z0-9][A-Z0-9][0-9][.-][0-9]+"/> <xs:pattern value="[A-NR-Z][0-9][A-Z][A-Z0-9][A-Z0-9][0-9][.-][0-9]+"/> <xs:pattern value="[A-NR-Z][0-9][A-Z][A-Z0-9][A-Z0-9][0-9][A-Z][A-Z0-9][A-Z0-9][0-9][.-][0-9]+"/> </xs:restriction> </xs:simpleType> |
UniprotAccession simpleType
| type | restriction of ExtendedUniprotAccession | |||||||||||||||
| facets |
|
|||||||||||||||
| documentation | UniProt accession number, without the sequence version and splice-variant suffix | |||||||||||||||
| source | <xs:simpleType name="UniprotAccession" sawsdl:modelReference="http://edamontology.org/data_1099"> <xs:annotation> <xs:documentation>UniProt accession number, without the sequence version and splice-variant suffix</xs:documentation> </xs:annotation> <xs:restriction base="ExtendedUniprotAccession"> <xs:pattern value="[OPQ][0-9][A-Z0-9][A-Z0-9][A-Z0-9][0-9]"/> <xs:pattern value="[A-NR-Z][0-9][A-Z][A-Z0-9][A-Z0-9][0-9]"/> <xs:pattern value="[A-NR-Z][0-9][A-Z][A-Z0-9][A-Z0-9][0-9][A-Z][A-Z0-9][A-Z0-9][0-9]"/> </xs:restriction> </xs:simpleType> |
ExtendedGenbankAccession simpleType
| type | restriction of Accession | ||||||||||||||||||||||||||||||
| used by |
|
||||||||||||||||||||||||||||||
| facets |
|
||||||||||||||||||||||||||||||
| source | <xs:simpleType name="ExtendedGenbankAccession"> <xs:restriction base="Accession"> <xs:pattern value="[A-Z][0-9]{5}(.[0-9]+)?"/> <xs:pattern value="[A-Z]{2}[0-9]{6}(.[0-9]+)?"/> <xs:pattern value="[A-Z]{3}[0-9]{5}(.[0-9]+)?"/> <xs:pattern value="[A-Z]{4}[0-9]{8,10}(.[0-9]+)?"/> <xs:pattern value="[A-Z]{5}[0-9]{7}(.[0-9]+)?"/> <xs:pattern value="[A-Z]{2}_[0-9]{6}(.[0-9]+)?"/> <xs:pattern value="[A-Z]{2}_[0-9]{9}(.[0-9]+)?"/> <xs:pattern value="[A-Z]{2}_[A-Z]{4}[0-9]{8,10}(.[0-9]+)?"/> </xs:restriction> </xs:simpleType> |
GenbankAccession simpleType
| type | restriction of ExtendedGenbankAccession | ||||||||||||||||||||||||||||||
| used by |
|
||||||||||||||||||||||||||||||
| facets |
|
||||||||||||||||||||||||||||||
| source | <xs:simpleType name="GenbankAccession"> <xs:restriction base="ExtendedGenbankAccession"> <xs:pattern value="[A-Z][0-9]{5}"/> <xs:pattern value="[A-Z]{2}[0-9]{6}"/> <xs:pattern value="[A-Z]{3}[0-9]{5}"/> <xs:pattern value="[A-Z]{4}[0-9]{8,10}"/> <xs:pattern value="[A-Z]{5}[0-9]{7}"/> <xs:pattern value="[A-Z]{2}_[0-9]{6}"/> <xs:pattern value="[A-Z]{2}_[0-9]{9}"/> <xs:pattern value="[A-Z]{2}_[A-Z]{4}[0-9]{8,10}"/> </xs:restriction> </xs:simpleType> |
GenbankNucleotideAccession simpleType
| type | restriction of GenbankAccession | ||||||||||||||||||
| facets |
|
||||||||||||||||||
| documentation | GenBank/EMBL/DDBJ nucleotide accession number | ||||||||||||||||||
| source | <xs:simpleType name="GenbankNucleotideAccession" sawsdl:modelReference="http://edamontology.org/data_1108"> <xs:annotation> <xs:documentation>GenBank/EMBL/DDBJ nucleotide accession number</xs:documentation> </xs:annotation> <xs:restriction base="GenbankAccession"> <xs:pattern value="[A-Z][0-9]{5}"/> <xs:pattern value="[A-Z]{2}[0-9]{6}"/> <xs:pattern value="[A-Z]{2}_[0-9]{6}"/> <xs:pattern value="[A-Z]{2}_[0-9]{9}"/> </xs:restriction> </xs:simpleType> |
GenbankProteinAccession simpleType
| type | restriction of GenbankAccession | |||||||||
| facets |
|
|||||||||
| documentation | GenBank/EMBL/DDBJ protein accession number | |||||||||
| source | <xs:simpleType name="GenbankProteinAccession"> <xs:annotation> <xs:documentation>GenBank/EMBL/DDBJ protein accession number</xs:documentation> </xs:annotation> <xs:restriction base="GenbankAccession"> <xs:pattern value="[A-Z]{3}[0-9]{5}"/> </xs:restriction> </xs:simpleType> |
GenbankWgsAccession simpleType
| type | restriction of GenbankAccession | ||||||||||||
| facets |
|
||||||||||||
| documentation | GenBank/EMBL/DDBJ WGS accession number | ||||||||||||
| source | <xs:simpleType name="GenbankWgsAccession"> <xs:annotation> <xs:documentation>GenBank/EMBL/DDBJ WGS accession number</xs:documentation> </xs:annotation> <xs:restriction base="GenbankAccession"> <xs:pattern value="[A-Z]{4}[0-9]{8,10}"/> <xs:pattern value="[A-Z]{2}_[A-Z]{4}[0-9]{8,10}"/> </xs:restriction> </xs:simpleType> |
GenbankMgaAccession simpleType
| type | restriction of GenbankAccession | |||||||||
| facets |
|
|||||||||
| documentation | GenBank/EMBL/DDBJ MGA accession number | |||||||||
| source | <xs:simpleType name="GenbankMgaAccession"> <xs:annotation> <xs:documentation>GenBank/EMBL/DDBJ MGA accession number</xs:documentation> </xs:annotation> <xs:restriction base="GenbankAccession"> <xs:pattern value="[A-Z]{5}[0-9]{7}"/> </xs:restriction> </xs:simpleType> |
PubmedId simpleType
| type | restriction of Accession | |||||||||
| facets |
|
|||||||||
| source | <xs:simpleType name="PubmedId"> <xs:restriction base="Accession"> <xs:pattern value="[1-9][0-9]{0,8}"/> </xs:restriction> </xs:simpleType> |
NcbiTaxonomyId simpleType
| type | restriction of Accession | |||||||||
| facets |
|
|||||||||
| documentation | NCBI Taxonomy ID (0 ... 999999999; i.e. a subset of 32-bit (4B) signed int) | |||||||||
| source | <xs:simpleType name="NcbiTaxonomyId" sawsdl:modelReference="http://edamontology.org/data_1179"> <xs:annotation> <xs:documentation>NCBI Taxonomy ID (0 ... 999999999; i.e. a subset of 32-bit (4B) signed int)</xs:documentation> </xs:annotation> <xs:restriction base="Accession"> <xs:pattern value="[1-9][0-9]{0,8}"/> </xs:restriction> </xs:simpleType> |
NcbiGeneticCodeId simpleType
| type | restriction of Accession | |||||||||
| facets |
|
|||||||||
| documentation | NCBI ID of a genetic code (1 ... 99) | |||||||||
| source | <xs:simpleType name="NcbiGeneticCodeId"> <xs:annotation> <xs:documentation>NCBI ID of a genetic code (1 ... 99)</xs:documentation> </xs:annotation> <xs:restriction base="Accession"> <xs:pattern value="[1-9][0-9]?"/> </xs:restriction> </xs:simpleType> |
RecommendedScoreName simpleType
| type | restriction of Name | |||||||||||||||||||||||||||||||||
| used by |
|
|||||||||||||||||||||||||||||||||
| facets |
|
|||||||||||||||||||||||||||||||||
| source | <xs:simpleType name="RecommendedScoreName"> <xs:restriction base="Name"> <xs:enumeration value="p-value" sawsdl:modelReference="http://edamontology.org/data_1669"/> <xs:enumeration value="E-value" sawsdl:modelReference="http://edamontology.org/data_1667"/> <xs:enumeration value="Standard deviation"/> <xs:enumeration value="Bit" sawsdl:modelReference="http://edamontology.org/data_2335"/> <xs:enumeration value="Z-score"/> <xs:enumeration value="Probability"/> <xs:enumeration value="Q3" sawsdl:modelReference="http://edamontology.org/NumberOfBlahblah"/> <xs:enumeration value="SOV" sawsdl:modelReference="http://edamontology.org/SegmentOverlap"/> </xs:restriction> </xs:simpleType> |
RecommendedVerdict simpleType
| type | restriction of Name | |||||||||||||||||||||
| used by |
|
|||||||||||||||||||||
| facets |
|
|||||||||||||||||||||
| documentation | Predefined, recommended verdict of a predicted or experimental evidence | |||||||||||||||||||||
| source | <xs:simpleType name="RecommendedVerdict"> <xs:annotation> <xs:documentation>Predefined, recommended verdict of a predicted or experimental evidence</xs:documentation> </xs:annotation> <xs:restriction base="Name"> <xs:enumeration value="Present"/> <xs:enumeration value="Putative"/> <xs:enumeration value="Improbable"/> <xs:enumeration value="Not present"/> </xs:restriction> </xs:simpleType> |
RecommendedReliability simpleType
| type | restriction of Name | |||||||||||||||
| used by |
|
|||||||||||||||
| facets |
|
|||||||||||||||
| documentation | Predefined, recommended reliability of an experimental evidence | |||||||||||||||
| source | <xs:simpleType name="RecommendedReliability"> <xs:annotation> <xs:documentation>Predefined, recommended reliability of an experimental evidence</xs:documentation> </xs:annotation> <xs:restriction base="Name"> <xs:enumeration value="Reliable"/> <xs:enumeration value="Disputable"/> </xs:restriction> </xs:simpleType> |
QualitativeCertainty simpleType
| type | restriction of Name | ||||||||||||||||||||||||||
| used by |
|
||||||||||||||||||||||||||
| facets |
|
||||||||||||||||||||||||||
| documentation | Qualitative certainty tag | ||||||||||||||||||||||||||
| source | <xs:simpleType name="QualitativeCertainty" sawsdl:modelReference="http://edamontology.org/format_2352"> <xs:annotation> <xs:documentation>Qualitative certainty tag</xs:documentation> </xs:annotation> <xs:restriction base="Name"> <xs:enumeration value="Certain"/> <xs:enumeration value="Uncertain"/> <xs:enumeration value="Value unknown"> <xs:annotation> <xs:documentation>The referred value completely unknown, not the certainty unknown</xs:documentation> </xs:annotation> </xs:enumeration> <xs:enumeration value="Certainly greater or equal"/> <xs:enumeration value="Certainly less or equal"/> </xs:restriction> </xs:simpleType> |
Strand simpleType
| type | restriction of Name | ||||||||||||||||||||||||
| used by |
|
||||||||||||||||||||||||
| facets |
|
||||||||||||||||||||||||
| source | <xs:simpleType name="Strand" sawsdl:modelReference="http://edamontology.org/data_0853 http://edamontology.org/format_2352"> <xs:restriction base="Name"> <xs:enumeration value="+"/> <xs:enumeration value="-"/> <xs:enumeration value="+ over start"/> <xs:enumeration value="- over start"/> <xs:enumeration value="Unknown"/> </xs:restriction> </xs:simpleType> |
OutsidePositionCorrespondance simpleType
| type | restriction of Name | ||||||||||||||||||||||||||||||||||
| used by |
|
||||||||||||||||||||||||||||||||||
| facets |
|
||||||||||||||||||||||||||||||||||
| documentation | Correspondence of the outside positions to the reference sequence | ||||||||||||||||||||||||||||||||||
| source | <xs:simpleType name="OutsidePositionCorrespondance" sawsdl:modelReference="http://edamontology.org/format_2352"> <xs:annotation> <xs:documentation>Correspondence of the outside positions to the reference sequence</xs:documentation> </xs:annotation> <xs:restriction base="Name"> <xs:enumeration value="In chromosome/scaffold/contig"> <xs:annotation> <xs:documentation>The outside position is in the nucleotide sequence of the either explicitly given chromosome or scaffold/contig, or the chromosome/scaffold/contig of the reference sequence. Outside-positions 1..m are positions in the chromosome</xs:documentation> </xs:annotation> </xs:enumeration> <xs:enumeration value="In explicit sequence"> <xs:annotation> <xs:documentation>The outside position is in an explicitly referenced nucleotide or amino-acid sequence. Can but does not have to be a supersequence of the reference sequence. Outside-positions 1..m are in the explicitly referenced supersequence. Fill in the 'explicitSequenceReference' element</xs:documentation> </xs:annotation> </xs:enumeration> <xs:enumeration value="In implicit supersequence"> <xs:annotation> <xs:documentation>The outside position is in a nucleotide or amino-acid supersequence (respectively) of the reference subsequence. Positions 1..n correspond, position -1 is the preceding (next point to the left) from 1</xs:documentation> </xs:annotation> </xs:enumeration> <xs:enumeration value="Relative to 1st translated base"> <xs:annotation> <xs:documentation>The outside position is in a nucleotide sequence. Position 1 corresponds to the 1st base of the 1st translated codon within the reference isoform, position -1 is the next to the left from 1 (NB. CDSs include the start and stop codons: do not use this correspondence option for outside features of CDSs)</xs:documentation> </xs:annotation> </xs:enumeration> <xs:enumeration value="Relative to 1st transcribed base"> <xs:annotation> <xs:documentation>The outside position is in a nucleotide sequence. Position 1 corresponds to the 1st base transcribed within the reference isoform, position -1 is the next to the left from 1</xs:documentation> </xs:annotation> </xs:enumeration> </xs:restriction> </xs:simpleType> |
RecommendedDatabaseName simpleType
| type | restriction of Name | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| used by |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| facets |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| documentation | Predefined, recommended database name of a public database | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| source | <xs:simpleType name="RecommendedDatabaseName"> <xs:annotation> <xs:documentation>Predefined, recommended database name of a public database</xs:documentation> </xs:annotation> <xs:restriction base="Name"> <xs:enumeration value="EMBL"/> <xs:enumeration value="EMBLCON"/> <xs:enumeration value="EMBLANN"/> <xs:enumeration value="EMBLCDS"/> <xs:enumeration value="EMBLSVA"/> <xs:enumeration value="Ensembl Gene"/> <xs:enumeration value="Ensembl Transcript"/> <xs:enumeration value="UniProt"/> <xs:enumeration value="UniProt/Swiss-Prot"/> <xs:enumeration value="UniProt/TrEMBL"/> <xs:enumeration value="UniSave"/> <xs:enumeration value="UniRef100"/> <xs:enumeration value="UniRef90"/> <xs:enumeration value="UniRef50"/> <xs:enumeration value="UniParc"/> <xs:enumeration value="IPI"/> <xs:enumeration value="RefSeq"/> <xs:enumeration value="InterPro"/> <xs:enumeration value="PDB"/> <xs:enumeration value="HGVbase"/> <xs:enumeration value="GenomeReviews"/> <xs:enumeration value="EPO Proteins"/> <xs:enumeration value="JPO Proteins"/> <xs:enumeration value="KIPO Proteins"/> <xs:enumeration value="USPO Proteins"/> <xs:enumeration value="PubMed"/> <xs:enumeration value="Medline"/> <xs:enumeration value="DDBJ/GenBank/EMBL"/> <xs:enumeration value="PDB"/> <xs:enumeration value="DAD"/> <xs:enumeration value="PRF"/> <xs:enumeration value="Patent"/> <xs:enumeration value="NCBI Taxonomy"/> <xs:enumeration value="NCBI Genetic Code"/> </xs:restriction> </xs:simpleType> |
RecommendedOntologyName simpleType
| type | restriction of Name | |||||||||||||||||||||
| used by |
|
|||||||||||||||||||||
| facets |
|
|||||||||||||||||||||
| documentation | Predefined, recommended name of a public ontology or controlled vocabulary | |||||||||||||||||||||
| source | <xs:simpleType name="RecommendedOntologyName"> <xs:annotation> <xs:documentation>Predefined, recommended name of a public ontology or controlled vocabulary</xs:documentation> </xs:annotation> <xs:restriction base="Name"> <xs:enumeration value="Gene Ontology"/> <xs:enumeration value="Sequence Ontology"/> <xs:enumeration value="ChEBI"/> <xs:enumeration value="EDAM"/> </xs:restriction> </xs:simpleType> |
Flag complexType
| diagram | |||
| used by |
|
||
| source | <xs:complexType name="Flag"/> |
AnyInteger simpleType
| type | xs:long | ||||
| used by |
|
||||
| documentation | Any integer number (~-9.2x10^18 ... ~9.2x10^18). Represented by a 64-bit (8B) signed long integer (long, long long, int64, same as xs:long, different from xs:integer) | ||||
| source | <xs:simpleType name="AnyInteger"> <xs:annotation> <xs:documentation>Any integer number (~-9.2x10^18 ... ~9.2x10^18). Represented by a 64-bit (8B) signed long integer (long, long long, int64, same as xs:long, different from xs:integer)</xs:documentation> </xs:annotation> <xs:restriction base="xs:long"/> </xs:simpleType> |
NonnegativeInteger simpleType
| type | restriction of AnyInteger | ||||||
| used by |
|
||||||
| facets |
|
||||||
| documentation | Non-negative integer number (0 ... ~9.2x10^18). Represented by a 64-bit (8B) signed long integer | ||||||
| source | <xs:simpleType name="NonnegativeInteger"> <xs:annotation> <xs:documentation>Non-negative integer number (0 ... ~9.2x10^18). Represented by a 64-bit (8B) signed long integer</xs:documentation> </xs:annotation> <xs:restriction base="AnyInteger"> <xs:minInclusive value="0"/> </xs:restriction> </xs:simpleType> |
NonzeroInteger simpleType
| type | restriction of AnyInteger | ||||||
| used by |
|
||||||
| facets |
|
||||||
| documentation | Non-zero integer number (~-9.2x10^18 ... -1, 1 ... ~9.2x10^18). Represented by a 64-bit (8B) signed long integer | ||||||
| source | <xs:simpleType name="NonzeroInteger"> <xs:annotation> <xs:documentation>Non-zero integer number (~-9.2x10^18 ... -1, 1 ... ~9.2x10^18). Represented by a 64-bit (8B) signed long integer</xs:documentation> </xs:annotation> <xs:restriction base="AnyInteger"> <xs:pattern value="[\-+]?[1-9][0-9]*"/> </xs:restriction> </xs:simpleType> |
InsertionInteger simpleType
| type | restriction of NonzeroInteger | |||||||||
| used by |
|
|||||||||
| facets |
|
|||||||||
| documentation | Insertion-point specific integer number (-1, 1 ... ~9.2x10^18). Represented by a 64-bit (8B) signed long integer | |||||||||
| source | <xs:simpleType name="InsertionInteger"> <xs:annotation> <xs:documentation>Insertion-point specific integer number (-1, 1 ... ~9.2x10^18). Represented by a 64-bit (8B) signed long integer</xs:documentation> </xs:annotation> <xs:restriction base="NonzeroInteger"> <xs:minInclusive value="-1"/> </xs:restriction> </xs:simpleType> |
PositiveInteger simpleType
| type | restriction of NonzeroInteger | |||||||||
| used by |
|
|||||||||
| facets |
|
|||||||||
| documentation | Positive integer number (1 ... ~9.2x10^18). Represented by a 64-bit (8B) signed long integer | |||||||||
| source | <xs:simpleType name="PositiveInteger"> <xs:annotation> <xs:documentation>Positive integer number (1 ... ~9.2x10^18). Represented by a 64-bit (8B) signed long integer</xs:documentation> </xs:annotation> <xs:restriction base="NonzeroInteger"> <xs:minInclusive value="1"/> </xs:restriction> </xs:simpleType> |
AnyDecimal simpleType
| type | xs:double | ||
| used by |
|
||
| documentation | Any decimal number with double precision (-INF, ~-1.7E308 ... 0, ~5E-324 subnormal ... ~2.3E-308 ... ~1.7E308, INF; +NaN). Represented by a 64-bit (8B) signed floating point number (double, long double, same as xs:double, different from xs:decimal) | ||
| source | <xs:simpleType name="AnyDecimal"> <xs:annotation> <xs:documentation>Any decimal number with double precision (-INF, ~-1.7E308 ... 0, ~5E-324 subnormal ... ~2.3E-308 ... ~1.7E308, INF; +NaN). Represented by a 64-bit (8B) signed floating point number (double, long double, same as xs:double, different from xs:decimal)</xs:documentation> </xs:annotation> <xs:restriction base="xs:double"/> </xs:simpleType> |
Probability simpleType
| type | restriction of AnyDecimal | |||||||||
| used by |
|
|||||||||
| facets |
|
|||||||||
| documentation | Probability with double precision (0 ... 1). Represented by a 64-bit (8B) signed floating point number | |||||||||
| source | <xs:simpleType name="Probability"> <xs:annotation> <xs:documentation>Probability with double precision (0 ... 1). Represented by a 64-bit (8B) signed floating point number</xs:documentation> </xs:annotation> <xs:restriction base="AnyDecimal"> <xs:minInclusive value="0"/> <xs:maxInclusive value="1"/> </xs:restriction> </xs:simpleType> |
Certainty simpleType
| type | union of (Probability, QualitativeCertainty) | ||
| used by |
|
||
| documentation | Quantitative or qualitative certainty | ||
| source | <xs:simpleType name="Certainty"> <xs:annotation> <xs:documentation>Quantitative or qualitative certainty</xs:documentation> </xs:annotation> <xs:union memberTypes="Probability QualitativeCertainty"/> </xs:simpleType> |
Verdict simpleType
| type | union of (Name, RecommendedVerdict) | ||
| used by |
|
||
| source | <xs:simpleType name="Verdict"> <xs:union memberTypes="Name RecommendedVerdict"/> </xs:simpleType> |
Reliability simpleType
| type | union of (Name, RecommendedReliability) | ||
| used by |
|
||
| source | <xs:simpleType name="Reliability"> <xs:union memberTypes="Name RecommendedReliability"/> </xs:simpleType> |
ScoreValue simpleType
| type | union of (AnyDecimal, Name, RecommendedVerdict, RecommendedReliability) | ||
| used by |
|
||
| source | <xs:simpleType name="ScoreValue"> <xs:union memberTypes="AnyDecimal Name RecommendedVerdict RecommendedReliability"/> </xs:simpleType> |
Uri simpleType
| type | restriction of xs:string | ||||||||||||||||||
| used by |
|
||||||||||||||||||
| facets |
|
||||||||||||||||||
| documentation | An absolute Unified Resource Identifier (URI), possibly a Web link. NB. Supports a subset of RFC 3986 generic syntax (selected schemes, DNS only, no user info, constrained port and characters) |
||||||||||||||||||
| source | <xs:simpleType name="Uri" sawsdl:modelReference="http://purl.org/dc/elements/1.1/identifier http://edamontology.org/data_0842 http://edamontology.org/data_1047 http://edamontology.org/format_2334"> <xs:annotation> <xs:documentation>An absolute Unified Resource Identifier (URI), possibly a Web link. NB. Supports a subset of RFC 3986 generic syntax (selected schemes, DNS only, no user info, constrained port and characters)</xs:documentation> </xs:annotation> <xs:restriction base="xs:string"> <xs:maxLength value="1000"/> <xs:pattern value="http://[a-z0-9]([a-z0-9]|\-[a-z0-9])*(\.[a-z0-9]([a-z0-9]|\-[a-z0-9])*)*(:[2-9][0-9]{3,})?(/([A-Za-z0-9\-._~!$'()*+,;=&]|%[0-9A-F]{2})+)*/?(\?([A-Za-z0-9\-._~!$'()*+,;=&/\?]|%[0-9A-F]{2})*)?(#([A-Za-z0-9\-._~!$'()*+,;=&/\?]|%[0-9A-F]{2})*)?"/> <xs:pattern value="https://[a-z0-9]([a-z0-9]|\-[a-z0-9])*(\.[a-z0-9]([a-z0-9]|\-[a-z0-9])*)*(:[2-9][0-9]{3,})?(/([A-Za-z0-9\-._~!$'()*+,;=&]|%[0-9A-F]{2})+)*/?(\?([A-Za-z0-9\-._~!$'()*+,;=&/\?]|%[0-9A-F]{2})*)?(#([A-Za-z0-9\-._~!$'()*+,;=&/\?]|%[0-9A-F]{2})*)?"/> <xs:pattern value="ftp://[a-z0-9]([a-z0-9]|\-[a-z0-9])*(\.[a-z0-9]([a-z0-9]|\-[a-z0-9])*)*(:[2-9][0-9]{3,})?(/([A-Za-z0-9\-._~!$'()*+,;=&]|%[0-9A-F]{2})+)*/?(\?([A-Za-z0-9\-._~!$'()*+,;=&/\?]|%[0-9A-F]{2})*)?(#([A-Za-z0-9\-._~!$'()*+,;=&/\?]|%[0-9A-F]{2})*)?"/> <xs:pattern value="urn:lsid:[a-z0-9]([a-z0-9]|\-[a-z0-9])*(\.[a-z0-9]([a-z0-9]|\-[a-z0-9])*)*:[A-Za-z0-9\-._~]+:[A-Za-z0-9\-._~]+(:[A-Za-z0-9\-._~]+)?"/> <!--= These URI patterns (regular expressions) are original work of the BioXSD editors, and thus - likewise the whole BioXSD data model and schema - are available under the restrictions of the Creative Commons Attribution-ShareAlike 4.0 International License (CC BY-SA 4.0). =--> </xs:restriction> <!--"Identifier"(0842)--> </xs:simpleType> |
Text simpleType
| type | restriction of xs:string | ||||||||||||
| used by |
|
||||||||||||
| facets |
|
||||||||||||
| documentation | Any plain text, possibly formatted. Can include new lines, tabs, or any number of consecutive spaces | ||||||||||||
| source | <xs:simpleType name="Text" sawsdl:modelReference="http://edamontology.org/data_0969 http://edamontology.org/format_2330"> <xs:annotation> <xs:documentation>Any plain text, possibly formatted. Can include new lines, tabs, or any number of consecutive spaces</xs:documentation> </xs:annotation> <xs:restriction base="xs:string"> <xs:minLength value="1"/> <xs:whiteSpace value="preserve"/> <xs:maxLength value="1000000"/> </xs:restriction> <!--"Free text"(0969)--> <!--"Text format"(2330)--> </xs:simpleType> |
Name simpleType
| type | restriction of xs:string | |||||||||
| used by |
|
|||||||||
| facets |
|
|||||||||
| source | <xs:simpleType name="Name" sawsdl:modelReference="http://www.w3.org/2000/01/rdf-schema#label http://edamontology.org/data_2099 http://edamontology.org/format_2330"> <xs:restriction base="xs:string"> <xs:maxLength value="1000"/> <xs:pattern value="\S(\S|\s\S)*"/> </xs:restriction> <!--= NB. Different from xs:Name which is a valid name of an XML tag. =--> </xs:simpleType> |
ChromosomeName simpleType
| type | restriction of Name | |||||||||||||||
| used by |
|
|||||||||||||||
| facets |
|
|||||||||||||||
| source | <xs:simpleType name="ChromosomeName" sawsdl:modelReference="http://edamontology.org/data_0987"> <xs:restriction base="Name"> <xs:pattern value="[A-Z][A-Za-z]*"/> <xs:pattern value="[0-9]+[A-Za-z]*"/> <xs:pattern value="[a-z]+[0-9]+[A-Za-z]*"/> </xs:restriction> </xs:simpleType> |
DatabaseName simpleType
| type | union of (Name, RecommendedDatabaseName) | ||
| used by |
|
||
| documentation | Name of a public or private database (predefined or arbitrary name) | ||
| source | <xs:simpleType name="DatabaseName" sawsdl:modelReference="http://www.w3.org/2000/01/rdf-schema#label http://edamontology.org/data_1056"> <xs:annotation> <xs:documentation>Name of a public or private database (predefined or arbitrary name)</xs:documentation> </xs:annotation> <xs:union memberTypes="Name RecommendedDatabaseName"/> <!--"Database name"(1056)--> </xs:simpleType> |
OntologyName simpleType
| type | union of (Name, RecommendedOntologyName) | ||
| used by |
|
||
| documentation | Name of a public or private ontology or controlled vocabulary (predefined or arbitrary name) | ||
| source | <xs:simpleType name="OntologyName" sawsdl:modelReference="http://edamontology.org/data_1051"> <xs:annotation> <xs:documentation>Name of a public or private ontology or controlled vocabulary (predefined or arbitrary name)</xs:documentation> </xs:annotation> <xs:union memberTypes="Name RecommendedOntologyName"/> </xs:simpleType> |
ScoreName simpleType
| type | union of (Name, RecommendedScoreName) | ||
| used by |
|
||
| source | <xs:simpleType name="ScoreName"> <xs:union memberTypes="Name RecommendedScoreName"/> </xs:simpleType> |
Phase simpleType
| type | restriction of NonnegativeInteger | |||||||||
| used by |
|
|||||||||
| facets |
|
|||||||||
| documentation | Phase of an incomplete peptide-coding nucleotide sequence, in the direction of translation | |||||||||
| source | <xs:simpleType name="Phase" sawsdl:modelReference="http://edamontology.org/data_2336"> <xs:annotation> <xs:documentation>Phase of an incomplete peptide-coding nucleotide sequence, in the direction of translation</xs:documentation> </xs:annotation> <xs:restriction base="NonnegativeInteger"> <xs:maxInclusive value="2"/> </xs:restriction> </xs:simpleType> |