| schema location: | http://bioxsd.org/BioXSD-1.0.xsd |
| attribute form default: | unqualified |
| element form default: | qualified |
| targetNamespace: | http://bioxsd.org/BioXSD-1.0 |
Empty complexType
| diagram | |||
| properties |
|
||
| used by | |||
| source | <xs:complexType name="Empty" final="#all"/> |
AnyInteger simpleType
| type | xs:long | ||||
| used by |
|
||||
| Documentation | Any integer number (~-9.2x10^18 ... ~9.2x10^18). Represented by a 64-bit (8B) signed long integer (long, long long, int64, same as xs:long, different from xs:integer) | ||||
| source | <xs:simpleType name="AnyInteger"> <xs:annotation> <xs:documentation> Any integer number (~-9.2x10^18 ... ~9.2x10^18). Represented by a 64-bit (8B) signed long integer (long, long long, int64, same as xs:long, different from xs:integer) </xs:documentation> </xs:annotation> <xs:restriction base="xs:long"/> </xs:simpleType> |
NonnegativeInteger simpleType
| type | restriction of AnyInteger | ||||||
| used by |
|
||||||
| facets |
|
||||||
| Documentation | Non-negative integer number (0 ... ~9.2x10^18). Represented by a 64-bit (8B) signed long integer | ||||||
| source | <xs:simpleType name="NonnegativeInteger"> <xs:annotation> <xs:documentation>Non-negative integer number (0 ... ~9.2x10^18). Represented by a 64-bit (8B) signed long integer</xs:documentation> </xs:annotation> <xs:restriction base="AnyInteger"> <xs:minInclusive value="0"/> </xs:restriction> </xs:simpleType> |
NonzeroInteger simpleType
| type | restriction of AnyInteger | ||||||
| used by |
|
||||||
| facets |
|
||||||
| Documentation | Non-zero integer number (~-9.2x10^18 ... -1, 1 ... ~9.2x10^18). Represented by a 64-bit (8B) signed long integer | ||||||
| source | <xs:simpleType name="NonzeroInteger"> <xs:annotation> <xs:documentation>Non-zero integer number (~-9.2x10^18 ... -1, 1 ... ~9.2x10^18). Represented by a 64-bit (8B) signed long integer</xs:documentation> </xs:annotation> <xs:restriction base="AnyInteger"> <xs:pattern value="[\-+]?[1-9][0-9]*"/> </xs:restriction> </xs:simpleType> |
InsertionInteger simpleType
| type | restriction of NonzeroInteger | |||||||||
| used by |
|
|||||||||
| facets |
|
|||||||||
| Documentation | Insertion-point specific integer number (-1, 1 ... ~9.2x10^18). Represented by a 64-bit (8B) signed long integer | |||||||||
| source | <xs:simpleType name="InsertionInteger"> <xs:annotation> <xs:documentation>Insertion-point specific integer number (-1, 1 ... ~9.2x10^18). Represented by a 64-bit (8B) signed long integer</xs:documentation> </xs:annotation> <xs:restriction base="NonzeroInteger"> <xs:minInclusive value="-1"/> </xs:restriction> </xs:simpleType> |
PositiveInteger simpleType
| type | restriction of NonzeroInteger | |||||||||
| used by |
|
|||||||||
| facets |
|
|||||||||
| Documentation | Positive integer number (1 ... ~9.2x10^18). Represented by a 64-bit (8B) signed long integer | |||||||||
| source | <xs:simpleType name="PositiveInteger"> <xs:annotation> <xs:documentation>Positive integer number (1 ... ~9.2x10^18). Represented by a 64-bit (8B) signed long integer</xs:documentation> </xs:annotation> <xs:restriction base="NonzeroInteger"> <xs:minInclusive value="1"/> </xs:restriction> </xs:simpleType> |
AnyDecimal simpleType
| type | xs:double | ||||
| used by |
|
||||
| Documentation | Any decimal number with double precision (-INF, ~-1.7E308 ... 0, ~5E-324 subnormal ... ~2.3E-308 ... ~1.7E308, INF; +NaN). Represented by a 64-bit (8B) signed floating point number (double, long double, same as xs:double, different from xs:decimal) | ||||
| source | <xs:simpleType name="AnyDecimal"> <xs:annotation> <xs:documentation> Any decimal number with double precision (-INF, ~-1.7E308 ... 0, ~5E-324 subnormal ... ~2.3E-308 ... ~1.7E308, INF; +NaN). Represented by a 64-bit (8B) signed floating point number (double, long double, same as xs:double, different from xs:decimal) </xs:documentation> </xs:annotation> <xs:restriction base="xs:double"/> </xs:simpleType> |
Probability simpleType
| type | restriction of AnyDecimal | |||||||||
| used by |
|
|||||||||
| facets |
|
|||||||||
| Documentation | Probability with double precision (0 ... 1). Represented by a 64-bit (8B) signed floating point number | |||||||||
| source | <xs:simpleType name="Probability"> <xs:annotation> <xs:documentation>Probability with double precision (0 ... 1). Represented by a 64-bit (8B) signed floating point number</xs:documentation> </xs:annotation> <xs:restriction base="AnyDecimal"> <xs:minInclusive value="0"/> <xs:maxInclusive value="1"/> </xs:restriction> </xs:simpleType> |
Certainty simpleType
| type | union of (Probability, QualitativeCertainty) |
| used by | |
| Documentation | Quantitave or qualitative certainty |
| source | <xs:simpleType name="Certainty"> <xs:annotation> <xs:documentation>Quantitave or qualitative certainty</xs:documentation> </xs:annotation> <xs:union memberTypes="Probability QualitativeCertainty"/> </xs:simpleType> |
Uri simpleType
| type | restriction of xs:string | |||||||||||||||
| used by |
|
|||||||||||||||
| facets |
|
|||||||||||||||
| Documentation | An absolute Unified Resource Identifier (URI), possibly a Web link.NB. Supports a subset of RFC 3986 generic syntax (selected schemes, DNS only, no user info, constrained port and characters) | |||||||||||||||
| source | <xs:simpleType name="Uri" sawsdl:modelReference="http://purl.org/edam/data/0001047"> <xs:annotation> <xs:documentation> An absolute Unified Resource Identifier (URI), possibly a Web link. NB. Supports a subset of RFC 3986 generic syntax (selected schemes, DNS only, no user info, constrained port and characters) </xs:documentation> </xs:annotation> <xs:restriction base="xs:string"> <xs:pattern value="http://[a-z0-9]([a-z0-9]|\-[a-z0-9])*(\.[a-z0-9]([a-z0-9]|\-[a-z0-9])*)*(:[2-9][0-9]{3,})?(/([A-Za-z0-9\-._~!$'()*+,;=]|%[0-9A-F]{2})+)*(\?([A-Za-z0-9\-._~!$'()*+,;=/\?]|%[0-9A-F]{2})*)?(#([A-Za-z0-9\-._~!$'()*+,;=/\?]|%[0-9A-F]{2})*)?"/> <xs:pattern value="https://[a-z0-9]([a-z0-9]|\-[a-z0-9])*(\.[a-z0-9]([a-z0-9]|\-[a-z0-9])*)*(:[2-9][0-9]{3,})?(/([A-Za-z0-9\-._~!$'()*+,;=]|%[0-9A-F]{2})+)*(\?([A-Za-z0-9\-._~!$'()*+,;=/\?]|%[0-9A-F]{2})*)?(#([A-Za-z0-9\-._~!$'()*+,;=/\?]|%[0-9A-F]{2})*)?"/> <xs:pattern value="ftp://[a-z0-9]([a-z0-9]|\-[a-z0-9])*(\.[a-z0-9]([a-z0-9]|\-[a-z0-9])*)*(:[2-9][0-9]{3,})?(/([A-Za-z0-9\-._~!$'()*+,;=]|%[0-9A-F]{2})+)*(\?([A-Za-z0-9\-._~!$'()*+,;=/\?]|%[0-9A-F]{2})*)?(#([A-Za-z0-9\-._~!$'()*+,;=/\?]|%[0-9A-F]{2})*)?"/> <xs:pattern value="urn:lsid:[a-z0-9]([a-z0-9]|\-[a-z0-9])*(\.[a-z0-9]([a-z0-9]|\-[a-z0-9])*)*:[A-Za-z0-9\-._~]+:[A-Za-z0-9\-._~]+(:[A-Za-z0-9\-._~]+)?"/> </xs:restriction> </xs:simpleType> |
FreeText simpleType
| type | restriction of xs:string | |||||||||
| used by | ||||||||||
| facets |
|
|||||||||
| Documentation | Any plain text (possibly formatted) | |||||||||
| source | <xs:simpleType name="FreeText" sawsdl:modelReference="http://purl.org/edam/data/0000969"> <xs:annotation> <xs:documentation>Any plain text (possibly formatted)</xs:documentation> </xs:annotation> <xs:restriction base="xs:string"> <xs:minLength value="1"/> <xs:whiteSpace value="preserve"/> </xs:restriction> </xs:simpleType> |
Name simpleType
ChromosomeName simpleType
| type | restriction of Name | |||||||||
| used by |
|
|||||||||
| facets |
|
|||||||||
| source | <xs:simpleType name="ChromosomeName"> <xs:restriction base="Name"> <xs:pattern value="[A-Z][A-Za-z]*"/> <xs:pattern value="[0-9]+[A-Za-z]*"/> </xs:restriction> </xs:simpleType> |
DatabaseName simpleType
| type | union of (Name, RecommendedDatabaseName) | ||
| used by |
|
||
| Documentation | Name of a public or private database (predefined or arbitrary name) | ||
| source | <xs:simpleType name="DatabaseName" sawsdl:modelReference="http://purl.org/edam/data/0001056"> <xs:annotation> <xs:documentation>Name of a public or private database (predefined or arbitrary name)</xs:documentation> </xs:annotation> <xs:union memberTypes="Name RecommendedDatabaseName"/> </xs:simpleType> |
OntologyName simpleType
| type | union of (Name, RecommendedOntologyName) | ||
| used by |
|
||
| Documentation | Name of a public or private controlled vocabulary/ontology (predefined or arbitrary name) | ||
| source | <xs:simpleType name="OntologyName" sawsdl:modelReference="http://purl.org/edam/data/0001051"> <xs:annotation> <xs:documentation>Name of a public or private controlled vocabulary/ontology (predefined or arbitrary name)</xs:documentation> </xs:annotation> <xs:union memberTypes="Name RecommendedOntologyName"/> </xs:simpleType> |
ScoreName simpleType
| type | union of (Name, RecommendedScoreName) | ||
| used by |
|
||
| source | <xs:simpleType name="ScoreName"> <xs:union memberTypes="Name RecommendedScoreName"/> </xs:simpleType> |
Verdict simpleType
| type | union of (Name, RecommendedVerdict) | ||
| used by |
|
||
| source | <xs:simpleType name="Verdict"> <xs:union memberTypes="Name RecommendedVerdict"/> </xs:simpleType> |
Reliability simpleType
| type | union of (Name, RecommendedReliability) | ||
| used by |
|
||
| source | <xs:simpleType name="Reliability"> <xs:union memberTypes="Name RecommendedReliability"/> </xs:simpleType> |
Phase simpleType
| type | restriction of NonnegativeInteger | |||||||||
| used by |
|
|||||||||
| facets |
|
|||||||||
| Documentation | Phase of an incomplete peptide-coding nucleotide sequence, in the direction of translation | |||||||||
| source | <xs:simpleType name="Phase"> <xs:annotation> <xs:documentation>Phase of an incomplete peptide-coding nucleotide sequence, in the direction of translation</xs:documentation> </xs:annotation> <xs:restriction base="NonnegativeInteger"> <xs:maxInclusive value="2"/> </xs:restriction> </xs:simpleType> |
GeneralNucleotideSequence simpleType
| type | restriction of xs:string | |||||||||
| used by |
|
|||||||||
| facets |
|
|||||||||
| Documentation | Nucleotide sequence in any letter case, possibly with ambiguous ("degenerate") bases | |||||||||
| source | <xs:simpleType name="GeneralNucleotideSequence" sawsdl:modelReference="http://purl.org/edam/data/0001210"> <xs:annotation> <xs:documentation>Nucleotide sequence in any letter case, possibly with ambiguous ("degenerate") bases</xs:documentation> </xs:annotation> <xs:restriction base="xs:string"> <xs:pattern value="[acgtrykmswbdhvn]+"/> <xs:pattern value="[acgurykmswbdhvn]+"/> </xs:restriction> </xs:simpleType> |
GeneralAminoacidSequence simpleType
| type | restriction of xs:string | ||||||
| used by |
|
||||||
| facets |
|
||||||
| Documentation | Amino-acid sequence in capital letters, possibly with ambiguous residues (Asx, Xle, Glx, Xaa/Unk) and additional residues (Pyl and Sec) | ||||||
| source | <xs:simpleType name="GeneralAminoacidSequence" sawsdl:modelReference="http://purl.org/edam/data/0001219"> <xs:annotation> <xs:documentation> Amino-acid sequence in capital letters, possibly with ambiguous residues (Asx, Xle, Glx, Xaa/Unk) and additional residues (Pyl and Sec) </xs:documentation> </xs:annotation> <xs:restriction base="xs:string"> <xs:pattern value="[ACDEFGHIKLMNPQRSTVWYBJZXUO]+"/> </xs:restriction> </xs:simpleType> |
Biosequence simpleType
| type | union of (GeneralNucleotideSequence, GeneralAminoacidSequence) | ||
| used by |
|
||
| Documentation | Nucleotide or amino-acid sequence in any letter case, possibly with ambiguous bases and residues | ||
| source | <xs:simpleType name="Biosequence"> <xs:annotation> <xs:documentation> Nucleotide or amino-acid sequence in any letter case, possibly with ambiguous bases and residues </xs:documentation> </xs:annotation> <xs:union memberTypes="GeneralNucleotideSequence GeneralAminoacidSequence"/> </xs:simpleType> |
NucleotideSequence simpleType
| type | restriction of GeneralNucleotideSequence | |||||||||
| used by |
|
|||||||||
| facets |
|
|||||||||
| Documentation | Nucleotide sequence in any letter case, without ambiguous ("degenerate") bases | |||||||||
| source | <xs:simpleType name="NucleotideSequence" sawsdl:modelReference="http://purl.org/edam/data/0001211"> <xs:annotation> <xs:documentation>Nucleotide sequence in any letter case, without ambiguous ("degenerate") bases</xs:documentation> </xs:annotation> <xs:restriction base="GeneralNucleotideSequence"> <xs:pattern value="[acgt]+"/> <xs:pattern value="[acgu]+"/> </xs:restriction> </xs:simpleType> |
AminoacidSequence simpleType
| type | restriction of GeneralAminoacidSequence | ||||||
| used by |
|
||||||
| facets |
|
||||||
| Documentation | Amino-acid sequence in capital letters, without ambiguous and additional residues (Pyl, Sec) | ||||||
| source | <xs:simpleType name="AminoacidSequence" sawsdl:modelReference="http://purl.org/edam/data/0001218"> <xs:annotation> <xs:documentation>Amino-acid sequence in capital letters, without ambiguous and additional residues (Pyl, Sec)</xs:documentation> </xs:annotation> <xs:restriction base="GeneralAminoacidSequence"> <xs:pattern value="[ACDEFGHIKLMNPQRSTVWY]+"/> </xs:restriction> </xs:simpleType> |
BiosequenceRecord complexType
| diagram | 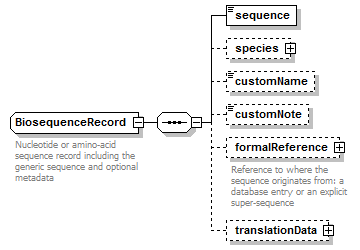 |
||||
| children | sequence species customName customNote formalReference translationData | ||||
| used by |
|
||||
| Documentation | Nucleotide or amino-acid sequence record including the generic sequence and optional metadata | ||||
| source | <xs:complexType name="BiosequenceRecord" sawsdl:modelReference="http://purl.org/edam/data/0000849"> <xs:annotation> <xs:documentation>Nucleotide or amino-acid sequence record including the generic sequence and optional metadata</xs:documentation> </xs:annotation> <xs:sequence> <xs:element name="sequence" type="Biosequence"/> <xs:element name="species" type="Species" minOccurs="0"/> <xs:element name="customName" type="Name" minOccurs="0"/> <xs:element name="customNote" type="FreeText" minOccurs="0"/> <xs:element name="formalReference" type="SequenceReference" minOccurs="0"> <xs:annotation> <xs:documentation>Reference to where the sequence originates from: a database entry or an explicit super-sequence</xs:documentation> </xs:annotation> </xs:element> <xs:element name="translationData" type="TranslationData" minOccurs="0"/> </xs:sequence> </xs:complexType> |
||||
| Model Reference | http://purl.org/edam/data/0000849 |
GeneralNucleotideSequenceRecord complexType
| diagram | 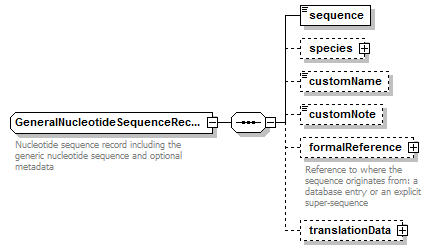 |
||||
| type | restriction of BiosequenceRecord | ||||
| properties |
|
||||
| children | sequence species customName customNote formalReference translationData | ||||
| used by |
|
||||
| Documentation | Nucleotide sequence record including the generic nucleotide sequence and optional metadata | ||||
| source | <xs:complexType name="GeneralNucleotideSequenceRecord" sawsdl:modelReference="http://purl.org/edam/data/0000849"> <xs:annotation> <xs:documentation>Nucleotide sequence record including the generic nucleotide sequence and optional metadata</xs:documentation> </xs:annotation> <xs:complexContent> <xs:restriction base="BiosequenceRecord"> <xs:sequence> <xs:element name="sequence" type="GeneralNucleotideSequence"/> <xs:element name="species" type="Species" minOccurs="0"/> <xs:element name="customName" type="Name" minOccurs="0"/> <xs:element name="customNote" type="FreeText" minOccurs="0"/> <xs:element name="formalReference" type="SequenceReference" minOccurs="0"> <xs:annotation> <xs:documentation>Reference to where the sequence originates from: a database entry or an explicit super-sequence</xs:documentation> </xs:annotation> </xs:element> <xs:element name="translationData" type="TranslationData" minOccurs="0"/> </xs:sequence> </xs:restriction> </xs:complexContent> </xs:complexType> |
||||
| Model Reference | http://purl.org/edam/data/0000849 |
GeneralAminoacidSequenceRecord complexType
| diagram | 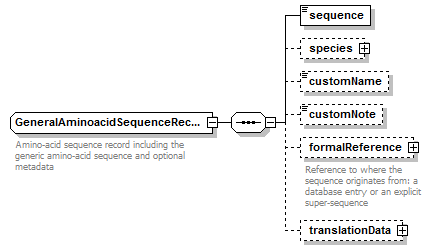 |
||||
| type | restriction of BiosequenceRecord | ||||
| properties |
|
||||
| children | sequence species customName customNote formalReference translationData | ||||
| used by |
|
||||
| Documentation | Amino-acid sequence record including the generic amino-acid sequence and optional metadata | ||||
| source | <xs:complexType name="GeneralAminoacidSequenceRecord" sawsdl:modelReference="http://purl.org/edam/data/0000849"> <xs:annotation> <xs:documentation>Amino-acid sequence record including the generic amino-acid sequence and optional metadata</xs:documentation> </xs:annotation> <xs:complexContent> <xs:restriction base="BiosequenceRecord"> <xs:sequence> <xs:element name="sequence" type="GeneralAminoacidSequence"/> <xs:element name="species" type="Species" minOccurs="0"/> <xs:element name="customName" type="Name" minOccurs="0"/> <xs:element name="customNote" type="FreeText" minOccurs="0"/> <xs:element name="formalReference" type="SequenceReference" minOccurs="0"> <xs:annotation> <xs:documentation>Reference to where the sequence originates from: a database entry or an explicit super-sequence</xs:documentation> </xs:annotation> </xs:element> <xs:element name="translationData" type="AminoacidTranslationData" minOccurs="0"/> </xs:sequence> </xs:restriction> </xs:complexContent> </xs:complexType> |
||||
| Model Reference | http://purl.org/edam/data/0000849 |
NucleotideSequenceRecord complexType
| diagram | 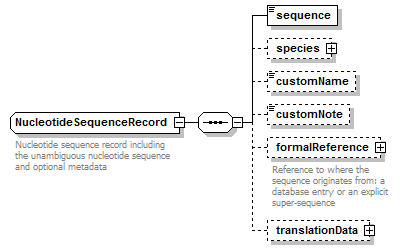 |
||
| type | restriction of GeneralNucleotideSequenceRecord | ||
| properties |
|
||
| children | sequence species customName customNote formalReference translationData | ||
| used by |
|
||
| Documentation | Nucleotide sequence record including the unambiguous nucleotide sequence and optional metadata | ||
| source | <xs:complexType name="NucleotideSequenceRecord" sawsdl:modelReference="http://purl.org/edam/data/0000849"> <xs:annotation> <xs:documentation>Nucleotide sequence record including the unambiguous nucleotide sequence and optional metadata</xs:documentation> </xs:annotation> <xs:complexContent> <xs:restriction base="GeneralNucleotideSequenceRecord"> <xs:sequence> <xs:element name="sequence" type="NucleotideSequence"/> <xs:element name="species" type="Species" minOccurs="0"/> <xs:element name="customName" type="Name" minOccurs="0"/> <xs:element name="customNote" type="FreeText" minOccurs="0"/> <xs:element name="formalReference" type="SequenceReference" minOccurs="0"> <xs:annotation> <xs:documentation>Reference to where the sequence originates from: a database entry or an explicit super-sequence</xs:documentation> </xs:annotation> </xs:element> <xs:element name="translationData" type="TranslationData" minOccurs="0"/> </xs:sequence> </xs:restriction> </xs:complexContent> </xs:complexType> |
||
| Model Reference | http://purl.org/edam/data/0000849 |
AminoacidSequenceRecord complexType
| diagram | 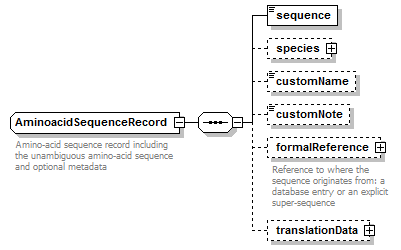 |
||
| type | restriction of GeneralAminoacidSequenceRecord | ||
| properties |
|
||
| children | sequence species customName customNote formalReference translationData | ||
| used by |
|
||
| Documentation | Amino-acid sequence record including the unambiguous amino-acid sequence and optional metadata | ||
| source | <xs:complexType name="AminoacidSequenceRecord" sawsdl:modelReference="http://purl.org/edam/data/0000849"> <xs:annotation> <xs:documentation>Amino-acid sequence record including the unambiguous amino-acid sequence and optional metadata</xs:documentation> </xs:annotation> <xs:complexContent> <xs:restriction base="GeneralAminoacidSequenceRecord"> <xs:sequence> <xs:element name="sequence" type="AminoacidSequence"/> <xs:element name="species" type="Species" minOccurs="0"/> <xs:element name="customName" type="Name" minOccurs="0"/> <xs:element name="customNote" type="FreeText" minOccurs="0"/> <xs:element name="formalReference" type="SequenceReference" minOccurs="0"> <xs:annotation> <xs:documentation>Reference to where the sequence originates from: a database entry or an explicit super-sequence</xs:documentation> </xs:annotation> </xs:element> <xs:element name="translationData" type="AminoacidTranslationData" minOccurs="0"/> </xs:sequence> </xs:restriction> </xs:complexContent> </xs:complexType> |
||
| Model Reference | http://purl.org/edam/data/0000849 |
GeneticCode complexType
| diagram | 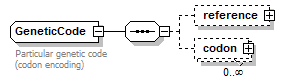 |
||
| children | reference codon | ||
| used by |
|
||
| Documentation | Particular genetic code (codon encoding) | ||
| source | <xs:complexType name="GeneticCode" sawsdl:modelReference="http://purl.org/edam/data/0001598"> <xs:annotation> <xs:documentation>Particular genetic code (codon encoding)</xs:documentation> </xs:annotation> <xs:sequence> <xs:element name="reference" type="EntryReference" minOccurs="0"/> <xs:element name="codon" minOccurs="0" maxOccurs="unbounded"> <xs:complexType> <xs:attribute name="code" use="required"> <xs:annotation> <xs:documentation>Codon code consisting of 3 bases (possibly ambiguous, "degenerate")</xs:documentation> </xs:annotation> <xs:simpleType> <xs:restriction base="GeneralNucleotideSequence"> <xs:length value="3"/> </xs:restriction> </xs:simpleType> </xs:attribute> <xs:attribute name="start" type="xs:boolean" default="false"/> <xs:attribute name="end" type="xs:boolean" default="false"/> <xs:attribute name="aminoacid"> <xs:annotation> <xs:documentation>One amino-acid (possibly ambiguous). NB. If the same codon codes for multiple amino-acids, use multiple 'codon' elements and fill in the 'note' attribute</xs:documentation> </xs:annotation> <xs:simpleType> <xs:restriction base="GeneralAminoacidSequence"> <xs:length value="1"/> </xs:restriction> </xs:simpleType> </xs:attribute> <xs:attribute name="note" type="FreeText"/> </xs:complexType> </xs:element> </xs:sequence> </xs:complexType> |
||
| Model Reference | http://purl.org/edam/data/0001598 |
TranslationData complexType
| diagram | 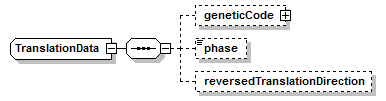 |
||||
| children | geneticCode phase reversedTranslationDirection | ||||
| used by |
|
||||
| source | <xs:complexType name="TranslationData"> <xs:sequence> <xs:element name="geneticCode" type="GeneticCode" minOccurs="0"/> <xs:element name="phase" type="Phase" minOccurs="0"/> <xs:element name="reversedTranslationDirection" type="Empty" minOccurs="0"/> </xs:sequence> </xs:complexType> |
AminoacidTranslationData complexType
| diagram | |||
| type | restriction of TranslationData | ||
| properties |
|
||
| children | geneticCode | ||
| used by |
|
||
| source | <xs:complexType name="AminoacidTranslationData"> <xs:complexContent> <xs:restriction base="TranslationData"> <xs:sequence> <xs:element name="geneticCode" type="GeneticCode" minOccurs="0"/> </xs:sequence> </xs:restriction> </xs:complexContent> </xs:complexType> |
DatabaseReference complexType
| diagram | 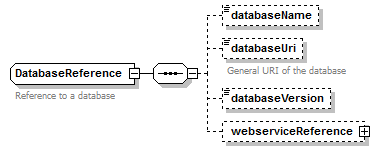 |
||
| children | databaseName databaseUri databaseVersion webserviceReference | ||
| used by |
|
||
| Documentation | Reference to a database | ||
| source | <xs:complexType name="DatabaseReference"> <xs:annotation> <xs:documentation>Reference to a database</xs:documentation> </xs:annotation> <xs:sequence> <xs:element name="databaseName" type="DatabaseName" minOccurs="0"/> <xs:element name="databaseUri" type="Uri" minOccurs="0"> <xs:annotation> <xs:documentation>General URI of the database</xs:documentation> </xs:annotation> </xs:element> <xs:element name="databaseVersion" type="Name" minOccurs="0" sawsdl:modelReference="http://purl.org/edam/data/0001670"/> <xs:element name="webserviceReference" type="WebserviceReference" minOccurs="0"/> </xs:sequence> </xs:complexType> |
EntryReference complexType
| diagram | 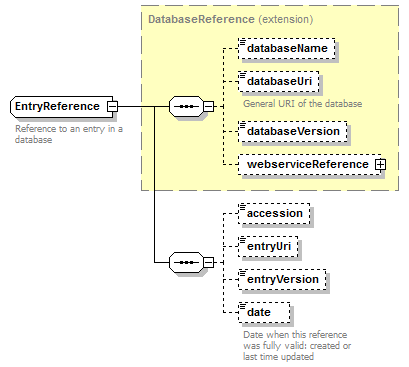 |
||||
| type | extension of DatabaseReference | ||||
| properties |
|
||||
| children | databaseName databaseUri databaseVersion webserviceReference accession entryUri entryVersion date | ||||
| used by |
|
||||
| Documentation | Reference to an entry in a database | ||||
| source | <xs:complexType name="EntryReference"> <xs:annotation> <xs:documentation>Reference to an entry in a database</xs:documentation> </xs:annotation> <xs:complexContent> <xs:extension base="DatabaseReference"> <xs:sequence> <xs:element name="accession" type="Accession" minOccurs="0"/> <xs:element name="entryUri" type="Uri" minOccurs="0" sawsdl:modelReference="http://purl.org/edam/data/0001054"/> <xs:element name="entryVersion" type="Name" minOccurs="0"/> <xs:element name="date" type="xs:date" minOccurs="0"> <xs:annotation> <xs:documentation>Date when this reference was fully valid: created or last time updated</xs:documentation> </xs:annotation> </xs:element> </xs:sequence> </xs:extension> </xs:complexContent> </xs:complexType> |
OntologyReference complexType
| diagram | 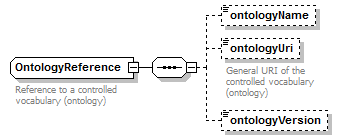 |
||
| children | ontologyName ontologyUri ontologyVersion | ||
| used by |
|
||
| Documentation | Reference to a controlled vocabulary (ontology) | ||
| source | <xs:complexType name="OntologyReference"> <xs:annotation> <xs:documentation>Reference to a controlled vocabulary (ontology)</xs:documentation> </xs:annotation> <xs:sequence> <xs:element name="ontologyName" type="OntologyName" minOccurs="0"/> <xs:element name="ontologyUri" type="Uri" minOccurs="0"> <xs:annotation> <xs:documentation>General URI of the controlled vocabulary (ontology)</xs:documentation> </xs:annotation> </xs:element> <xs:element name="ontologyVersion" type="Name" minOccurs="0"/> </xs:sequence> </xs:complexType> |
OntologyTerm complexType
| diagram | 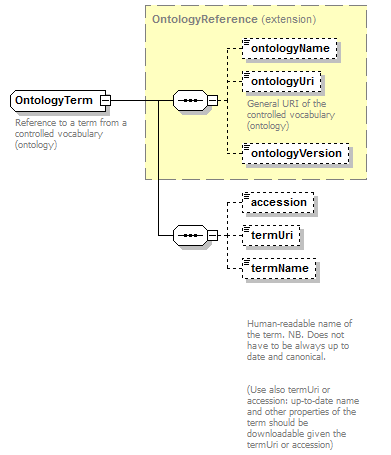 |
||
| type | extension of OntologyReference | ||
| properties |
|
||
| children | ontologyName ontologyUri ontologyVersion accession termUri termName | ||
| used by |
|
||
| Documentation | Reference to a term from a controlled vocabulary (ontology) | ||
| source | <xs:complexType name="OntologyTerm"> <xs:annotation> <xs:documentation>Reference to a term from a controlled vocabulary (ontology)</xs:documentation> </xs:annotation> <xs:complexContent> <xs:extension base="OntologyReference"> <xs:sequence> <xs:element name="accession" type="Accession" minOccurs="0" sawsdl:modelReference="http://purl.org/edam/data/0001087"/> <xs:element name="termUri" type="Uri" minOccurs="0" sawsdl:modelReference="http://purl.org/edam/data/0001087"/> <xs:element name="termName" type="Name" minOccurs="0"> <xs:annotation> <xs:documentation> Human-readable name of the term. NB. Does not have to be always up to date and canonical. (Use also termUri or accession: up-to-date name and other properties of the term should be downloadable given the termUri or accession)</xs:documentation> </xs:annotation> </xs:element> </xs:sequence> </xs:extension> </xs:complexContent> </xs:complexType> |
Species complexType
| diagram | 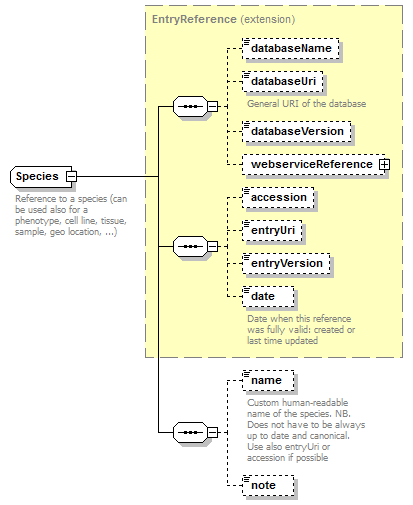 |
||
| type | extension of EntryReference | ||
| properties |
|
||
| children | databaseName databaseUri databaseVersion webserviceReference accession entryUri entryVersion date name note | ||
| used by |
|
||
| Documentation | Reference to a species (can be used also for a phenotype, cell line, tissue, sample, geo location, ...) | ||
| source | <xs:complexType name="Species"> <xs:annotation> <xs:documentation>Reference to a species (can be used also for a phenotype, cell line, tissue, sample, geo location, ...)</xs:documentation> </xs:annotation> <xs:complexContent> <xs:extension base="EntryReference"> <xs:sequence> <xs:element name="name" type="Name" minOccurs="0" sawsdl:modelReference="http://purl.org/edam/data/0001045"> <xs:annotation> <xs:documentation>Custom human-readable name of the species. NB. Does not have to be always up to date and canonical. Use also entryUri or accession if possible</xs:documentation> </xs:annotation> </xs:element> <xs:element name="note" type="FreeText" minOccurs="0"/> </xs:sequence> </xs:extension> </xs:complexContent> </xs:complexType> |
SequenceReference complexType
| diagram | 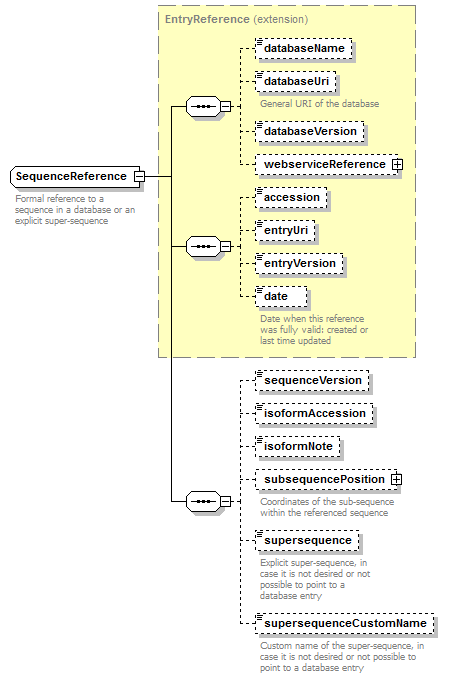 |
||
| type | extension of EntryReference | ||
| properties |
|
||
| children | databaseName databaseUri databaseVersion webserviceReference accession entryUri entryVersion date sequenceVersion isoformAccession isoformNote subsequencePosition supersequence supersequenceCustomName | ||
| used by |
|
||
| Documentation | Formal reference to a sequence in a database or an explicit super-sequence | ||
| source | <xs:complexType name="SequenceReference"> <xs:annotation> <xs:documentation>Formal reference to a sequence in a database or an explicit super-sequence</xs:documentation> </xs:annotation> <xs:complexContent> <xs:extension base="EntryReference"> <xs:sequence> <xs:element name="sequenceVersion" type="Name" minOccurs="0" sawsdl:modelReference="http://purl.org/edam/data/0001771"/> <xs:element name="isoformAccession" type="Accession" minOccurs="0"/> <xs:element name="isoformNote" type="Name" minOccurs="0"/> <xs:element name="subsequencePosition" type="SequencePosition" minOccurs="0"> <xs:annotation> <xs:documentation>Coordinates of the sub-sequence within the referenced sequence</xs:documentation> </xs:annotation> </xs:element> <xs:element name="supersequence" type="Biosequence" minOccurs="0"> <xs:annotation> <xs:documentation>Explicit super-sequence, in case it is not desired or not possible to point to a database entry</xs:documentation> </xs:annotation> </xs:element> <xs:element name="supersequenceCustomName" type="Name" minOccurs="0"> <xs:annotation> <xs:documentation>Custom name of the super-sequence, in case it is not desired or not possible to point to a database entry</xs:documentation> </xs:annotation> </xs:element> </xs:sequence> </xs:extension> </xs:complexContent> </xs:complexType> |
Method complexType
| diagram | 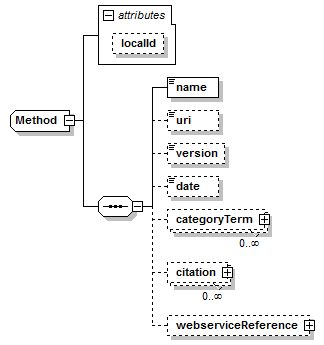 |
||||||||||||
| children | name uri version date categoryTerm citation webserviceReference | ||||||||||||
| used by |
|
||||||||||||
| attributes |
|
||||||||||||
| source | <xs:complexType name="Method"> <xs:sequence> <xs:element name="name" type="Name"/> <xs:element name="uri" type="Uri" minOccurs="0" sawsdl:modelReference="http://purl.org/edam/data/0000977"/> <xs:element name="version" type="Name" minOccurs="0" sawsdl:modelReference="http://purl.org/edam/data/0001671"/> <xs:element name="date" type="xs:date" minOccurs="0"/> <xs:element name="categoryTerm" type="OntologyTerm" minOccurs="0" maxOccurs="unbounded"/> <xs:element name="citation" type="EntryReference" minOccurs="0" maxOccurs="unbounded" sawsdl:modelReference="http://purl.org/edam/data/0000970"/> <xs:element name="webserviceReference" type="WebserviceReference" minOccurs="0"/> </xs:sequence> <xs:attribute name="localId" type="LocalReference"/> </xs:complexType> |
WebserviceReference complexType
| diagram | 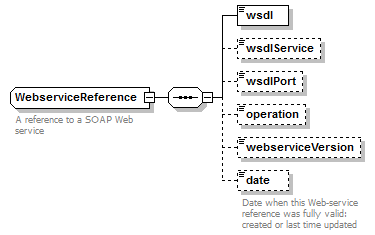 |
||
| children | wsdl wsdlService wsdlPort operation webserviceVersion date | ||
| used by |
|
||
| Documentation | A reference to a SOAP Web service | ||
| source | <xs:complexType name="WebserviceReference"> <xs:annotation> <xs:documentation>A reference to a SOAP Web service</xs:documentation> </xs:annotation> <xs:sequence> <xs:element name="wsdl" type="Uri"/> <xs:element name="wsdlService" type="xs:NMTOKEN" minOccurs="0"/> <xs:element name="wsdlPort" type="xs:NMTOKEN" minOccurs="0"/> <xs:element name="operation" type="xs:NMTOKEN" minOccurs="0"/> <xs:element name="webserviceVersion" type="Name" minOccurs="0"/> <xs:element name="date" type="xs:date" minOccurs="0"> <xs:annotation> <xs:documentation>Date when this Web-service reference was fully valid: created or last time updated</xs:documentation> </xs:annotation> </xs:element> </xs:sequence> </xs:complexType> |
LocalReference simpleType
| type | restriction of xs:string | ||||||
| used by |
|
||||||
| facets |
|
||||||
| Documentation | Identifier for local references within a data record | ||||||
| source | <xs:simpleType name="LocalReference"> <xs:annotation> <xs:documentation>Identifier for local references within a data record</xs:documentation> </xs:annotation> <xs:restriction base="xs:string"> <xs:pattern value="[A-Za-z0-9\-._~]+"/> </xs:restriction> </xs:simpleType> |
Accession simpleType
| type | restriction of xs:string | ||||||
| used by |
|
||||||
| facets |
|
||||||
| Documentation | Generalisation of bioinformatic accession numbers (stable primary keys/identifiers) | ||||||
| source | <xs:simpleType name="Accession" sawsdl:modelReference="http://purl.org/edam/data/0000976"> <xs:annotation> <xs:documentation>Generalisation of bioinformatic accession numbers (stable primary keys/identifiers)</xs:documentation> </xs:annotation> <xs:restriction base="xs:string"> <xs:pattern value="[A-Za-z0-9\-._~]+"/> </xs:restriction> </xs:simpleType> |
EmblAccession simpleType
| type | Accession | ||||||
| facets |
|
||||||
| source | <xs:simpleType name="EmblAccession" sawsdl:modelReference="http://purl.org/edam/data/0001107"> <xs:restriction base="Accession"/> </xs:simpleType> |
ExtendedUniprotAccession simpleType
| type | restriction of Accession | |||||||||||||||||||||
| used by |
|
|||||||||||||||||||||
| facets |
|
|||||||||||||||||||||
| Documentation | UniProt accession number, optionally with the sequence version or the splice-variant suffix | |||||||||||||||||||||
| source | <xs:simpleType name="ExtendedUniprotAccession"> <xs:annotation> <xs:documentation>UniProt accession number, optionally with the sequence version or the splice-variant suffix</xs:documentation> </xs:annotation> <xs:restriction base="Accession"> <xs:pattern value="[A-NR-Z][0-9][A-Z][A-Z0-9][A-Z0-9][0-9]"/> <xs:pattern value="[OPQ][0-9][A-Z0-9][A-Z0-9][A-Z0-9][0-9]"/> <xs:pattern value="[A-NR-Z][0-9][A-Z][A-Z0-9][A-Z0-9][0-9].[0-9]+"/> <xs:pattern value="[OPQ][0-9][A-Z0-9][A-Z0-9][A-Z0-9][0-9].[0-9]+"/> <xs:pattern value="[A-NR-Z][0-9][A-Z][A-Z0-9][A-Z0-9][0-9]-[0-9]+"/> <xs:pattern value="[OPQ][0-9][A-Z0-9][A-Z0-9][A-Z0-9][0-9]-[0-9]+"/> </xs:restriction> </xs:simpleType> |
UniprotAccession simpleType
| type | restriction of ExtendedUniprotAccession | |||||||||
| facets |
|
|||||||||
| Documentation | UniProt accession number, without the sequence version and splice-variant suffix | |||||||||
| source | <xs:simpleType name="UniprotAccession" sawsdl:modelReference="http://purl.org/edam/data/0001099"> <xs:annotation> <xs:documentation>UniProt accession number, without the sequence version and splice-variant suffix</xs:documentation> </xs:annotation> <xs:restriction base="ExtendedUniprotAccession"> <xs:pattern value="[A-NR-Z][0-9][A-Z][A-Z0-9][A-Z0-9][0-9]"/> <xs:pattern value="[OPQ][0-9][A-Z0-9][A-Z0-9][A-Z0-9][0-9]"/> </xs:restriction> </xs:simpleType> |
ExtendedGenbankAccession simpleType
| type | restriction of Accession | |||||||||||||||||||||||||||
| used by |
|
|||||||||||||||||||||||||||
| facets |
|
|||||||||||||||||||||||||||
| source | <xs:simpleType name="ExtendedGenbankAccession"> <xs:restriction base="Accession"> <xs:pattern value="[A-Z][0-9]{5}(.[0-9]+)?"/> <xs:pattern value="[A-Z]{2}[0-9]{6}(.[0-9]+)?"/> <xs:pattern value="[A-Z]{3}[0-9]{5}(.[0-9]+)?"/> <xs:pattern value="[A-Z]{4}[0-9]{8,10}(.[0-9]+)?"/> <xs:pattern value="[A-Z]{5}[0-9]{7}(.[0-9]+)?"/> <xs:pattern value="[A-Z]{2}_[0-9]{6}(.[0-9]+)?"/> <xs:pattern value="[A-Z]{2}_[0-9]{9}(.[0-9]+)?"/> <xs:pattern value="[A-Z]{2}_[A-Z]{4}[0-9]{8,10}(.[0-9]+)?"/> </xs:restriction> </xs:simpleType> |
GenbankAccession simpleType
| type | restriction of ExtendedGenbankAccession | |||||||||||||||||||||||||||
| used by |
|
|||||||||||||||||||||||||||
| facets |
|
|||||||||||||||||||||||||||
| source | <xs:simpleType name="GenbankAccession"> <xs:restriction base="ExtendedGenbankAccession"> <xs:pattern value="[A-Z][0-9]{5}"/> <xs:pattern value="[A-Z]{2}[0-9]{6}"/> <xs:pattern value="[A-Z]{3}[0-9]{5}"/> <xs:pattern value="[A-Z]{4}[0-9]{8,10}"/> <xs:pattern value="[A-Z]{5}[0-9]{7}"/> <xs:pattern value="[A-Z]{2}_[0-9]{6}"/> <xs:pattern value="[A-Z]{2}_[0-9]{9}"/> <xs:pattern value="[A-Z]{2}_[A-Z]{4}[0-9]{8,10}"/> </xs:restriction> </xs:simpleType> |
GenbankNucleotideAccession simpleType
| type | restriction of GenbankAccession | |||||||||||||||
| facets |
|
|||||||||||||||
| Documentation | GenBank/EMBL/DDBJ nucleotide accession number | |||||||||||||||
| source | <xs:simpleType name="GenbankNucleotideAccession" sawsdl:modelReference="http://purl.org/edam/data/0001108"> <xs:annotation> <xs:documentation>GenBank/EMBL/DDBJ nucleotide accession number</xs:documentation> </xs:annotation> <xs:restriction base="GenbankAccession"> <xs:pattern value="[A-Z][0-9]{5}"/> <xs:pattern value="[A-Z]{2}[0-9]{6}"/> <xs:pattern value="[A-Z]{2}_[0-9]{6}"/> <xs:pattern value="[A-Z]{2}_[0-9]{9}"/> </xs:restriction> </xs:simpleType> |
GenbankProteinAccession simpleType
| type | restriction of GenbankAccession | ||||||
| facets |
|
||||||
| Documentation | GenBank/EMBL/DDBJ protein accession number | ||||||
| source | <xs:simpleType name="GenbankProteinAccession"> <xs:annotation> <xs:documentation>GenBank/EMBL/DDBJ protein accession number</xs:documentation> </xs:annotation> <xs:restriction base="GenbankAccession"> <xs:pattern value="[A-Z]{3}[0-9]{5}"/> </xs:restriction> </xs:simpleType> |
GenbankWgsAccession simpleType
| type | restriction of GenbankAccession | |||||||||
| facets |
|
|||||||||
| Documentation | GenBank/EMBL/DDBJ WGS accession number | |||||||||
| source | <xs:simpleType name="GenbankWgsAccession"> <xs:annotation> <xs:documentation>GenBank/EMBL/DDBJ WGS accession number</xs:documentation> </xs:annotation> <xs:restriction base="GenbankAccession"> <xs:pattern value="[A-Z]{4}[0-9]{8,10}"/> <xs:pattern value="[A-Z]{2}_[A-Z]{4}[0-9]{8,10}"/> </xs:restriction> </xs:simpleType> |
GenbankMgaAccession simpleType
| type | restriction of GenbankAccession | ||||||
| facets |
|
||||||
| Documentation | GenBank/EMBL/DDBJ MGA accession number | ||||||
| source | <xs:simpleType name="GenbankMgaAccession"> <xs:annotation> <xs:documentation>GenBank/EMBL/DDBJ MGA accession number</xs:documentation> </xs:annotation> <xs:restriction base="GenbankAccession"> <xs:pattern value="[A-Z]{5}[0-9]{7}"/> </xs:restriction> </xs:simpleType> |
PubmedId simpleType
| type | restriction of Accession | ||||||
| facets |
|
||||||
| source | <xs:simpleType name="PubmedId"> <xs:restriction base="Accession"> <xs:pattern value="[0-9]{1,9}"/> </xs:restriction> </xs:simpleType> |
NcbiTaxonomyId simpleType
| type | restriction of Accession | ||||||
| facets |
|
||||||
| Documentation | NCBI Taxonomy ID (0 ... 999999999; i.e. a subset of 32-bit (4B) signed int) | ||||||
| source | <xs:simpleType name="NcbiTaxonomyId" sawsdl:modelReference="http://purl.org/edam/data/0001179"> <xs:annotation> <xs:documentation>NCBI Taxonomy ID (0 ... 999999999; i.e. a subset of 32-bit (4B) signed int)</xs:documentation> </xs:annotation> <xs:restriction base="Accession"> <xs:pattern value="[0-9]{1,9}"/> </xs:restriction> </xs:simpleType> |
NcbiGeneticCodeId simpleType
| type | restriction of Accession | ||||||
| facets |
|
||||||
| Documentation | NCBI ID of a genetic code (1 ... 99) | ||||||
| source | <xs:simpleType name="NcbiGeneticCodeId"> <xs:annotation> <xs:documentation>NCBI ID of a genetic code (1 ... 99)</xs:documentation> </xs:annotation> <xs:restriction base="Accession"> <xs:pattern value="[1-9][0-9]?"/> </xs:restriction> </xs:simpleType> |
OboTermId simpleType
| type | restriction of Accession | ||||||
| facets |
|
||||||
| Documentation | Term ID in an OBO-Foundry ontology | ||||||
| source | <xs:simpleType name="OboTermId" sawsdl:modelReference="http://purl.org/edam/data/0001087"> <xs:annotation> <xs:documentation>Term ID in an OBO-Foundry ontology</xs:documentation> </xs:annotation> <xs:restriction base="Accession"> <xs:pattern value="[0-9]{6,7}"/> </xs:restriction> </xs:simpleType> |
Feature complexType
| diagram | 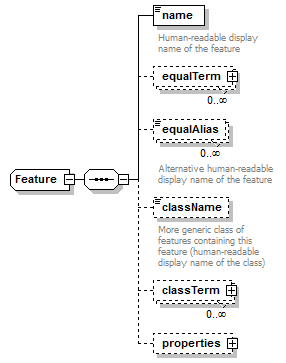 |
||
| children | name equalTerm equalAlias className classTerm properties | ||
| used by |
|
||
| source | <xs:complexType name="Feature"> <xs:sequence> <xs:element name="name" type="Name"> <xs:annotation> <xs:documentation>Human-readable display name of the feature</xs:documentation> </xs:annotation> </xs:element> <xs:element name="equalTerm" type="OntologyTerm" minOccurs="0" maxOccurs="unbounded"/> <xs:element name="equalAlias" type="Name" minOccurs="0" maxOccurs="unbounded"> <xs:annotation> <xs:documentation>Alternative human-readable display name of the feature</xs:documentation> </xs:annotation> </xs:element> <xs:element name="className" type="Name" minOccurs="0"> <xs:annotation> <xs:documentation>More generic class of features containing this feature (human-readable display name of the class)</xs:documentation> </xs:annotation> </xs:element> <xs:element name="classTerm" type="OntologyTerm" minOccurs="0" maxOccurs="unbounded"/> <xs:element name="properties" type="FeatureData" minOccurs="0"/> </xs:sequence> </xs:complexType> |
FeatureData complexType
| diagram | 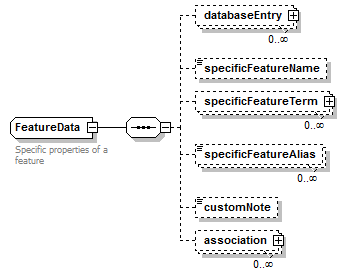 |
||
| children | databaseEntry specificFeatureName specificFeatureTerm specificFeatureAlias customNote association | ||
| used by |
|
||
| Documentation | Specific properties of a feature | ||
| source | <xs:complexType name="FeatureData"> <xs:annotation> <xs:documentation>Specific properties of a feature</xs:documentation> </xs:annotation> <xs:sequence> <xs:element name="databaseEntry" type="EntryReference" minOccurs="0" maxOccurs="unbounded"/> <xs:element name="specificFeatureName" type="Name" minOccurs="0"/> <xs:element name="specificFeatureTerm" type="OntologyTerm" minOccurs="0" maxOccurs="unbounded"/> <xs:element name="specificFeatureAlias" type="Name" minOccurs="0" maxOccurs="unbounded"/> <xs:element name="customNote" type="FreeText" minOccurs="0"/> <xs:element name="association" minOccurs="0" maxOccurs="unbounded"> <xs:complexType> <xs:sequence> <xs:element name="relationName" type="Name" minOccurs="0"/> <xs:element name="relationTerm" type="OntologyTerm" minOccurs="0" maxOccurs="unbounded"/> <xs:choice> <xs:element name="associatedFeature" type="Feature" maxOccurs="unbounded"/> <xs:element name="associatedTerm" type="OntologyTerm" maxOccurs="unbounded"/> <xs:element name="associatedDatabaseEntry" type="EntryReference" maxOccurs="unbounded"/> </xs:choice> </xs:sequence> </xs:complexType> </xs:element> </xs:sequence> </xs:complexType> |
FeatureOccurence complexType
| diagram | 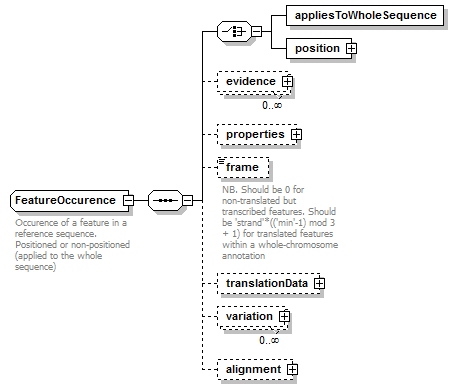 |
||
| children | appliesToWholeSequence position evidence properties frame translationData variation alignment | ||
| used by |
|
||
| Documentation | Occurence of a feature in a reference sequence. Positioned or non-positioned (applied to the whole sequence) | ||
| source | <xs:complexType name="FeatureOccurence"> <xs:annotation> <xs:documentation>Occurence of a feature in a reference sequence. Positioned or non-positioned (applied to the whole sequence)</xs:documentation> </xs:annotation> <xs:sequence> <xs:choice> <xs:element name="appliesToWholeSequence" type="Empty"/> <xs:element name="position" type="GeneralSequencePosition"/> </xs:choice> <xs:element name="evidence" type="Evidence" minOccurs="0" maxOccurs="unbounded"/> <xs:element name="properties" type="FeatureData" minOccurs="0"/> <xs:element name="frame" minOccurs="0"> <xs:annotation> <xs:documentation>NB. Should be 0 for non-translated but transcribed features. Should be 'strand'*(('min'-1) mod 3 + 1) for translated features within a whole-chromosome annotation</xs:documentation> </xs:annotation> <xs:simpleType> <xs:restriction base="AnyInteger"> <xs:minInclusive value="-3"/> <xs:maxInclusive value="3"/> </xs:restriction> </xs:simpleType> </xs:element> <xs:element name="translationData" type="TranslationData" minOccurs="0"/> <xs:element name="variation" minOccurs="0" maxOccurs="unbounded"> <xs:complexType> <xs:sequence> <xs:element name="original" type="Biosequence" minOccurs="0" maxOccurs="unbounded"/> <xs:choice maxOccurs="unbounded"> <xs:element name="variant" type="Biosequence"/> <xs:element name="missing" type="Empty"/> </xs:choice> <xs:element name="position" type="SequencePosition" minOccurs="0"> <xs:annotation> <xs:documentation>NB. Corresponds to a position in the reference sequence (not to a position within the feature occurence)</xs:documentation> </xs:annotation> </xs:element> </xs:sequence> </xs:complexType> </xs:element> <xs:element name="alignment" minOccurs="0" sawsdl:modelReference="http://purl.org/edam/data/0001381"> <xs:complexType> <xs:sequence> <xs:element name="alignmentOfReference" minOccurs="0"> <xs:complexType> <xs:sequence> <xs:element name="gap" type="Gap" minOccurs="0" maxOccurs="unbounded"/> <xs:element name="frameshift" type="Frameshift" minOccurs="0" maxOccurs="unbounded"/> <xs:element name="sequenceScore" type="Result" minOccurs="0" maxOccurs="unbounded"/> </xs:sequence> </xs:complexType> <xs:unique name="uniqueGapStart_ar"> <xs:selector xpath="gap"/> <xs:field xpath="@start"/> </xs:unique> <xs:unique name="uniqueFrameshiftPos_ar"> <xs:selector xpath="frameshift"/> <xs:field xpath="@pos"/> </xs:unique> </xs:element> <xs:element name="alignedSequence" type="AlignedBiosequence"> <xs:unique name="uniqueGapStart_as"> <xs:selector xpath="gap"/> <xs:field xpath="@start"/> </xs:unique> <xs:unique name="uniqueFrameshiftPos_as"> <xs:selector xpath="frameshift"/> <xs:field xpath="@pos"/> </xs:unique> </xs:element> </xs:sequence> </xs:complexType> </xs:element> </xs:sequence> </xs:complexType> |
AnnotatedSequence complexType
| diagram | 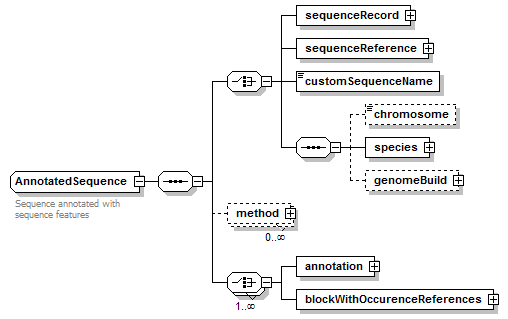 |
| children | sequenceRecord sequenceReference customSequenceName chromosome species genomeBuild method annotation blockWithOccurenceReferences |
| Documentation | Sequence annotated with sequence features |
| source | <xs:complexType name="AnnotatedSequence" sawsdl:modelReference="http://purl.org/edam/data/0001255"> <xs:annotation> <xs:documentation>Sequence annotated with sequence features</xs:documentation> </xs:annotation> <xs:sequence> <!--= Reference sequence =--> <xs:choice> <xs:element name="sequenceRecord" type="BiosequenceRecord"/> <xs:element name="sequenceReference" type="SequenceReference"/> <xs:element name="customSequenceName" type="Name"/> <xs:sequence> <xs:element name="chromosome" type="ChromosomeName" minOccurs="0"/> <xs:element name="species" type="Species"/> <xs:element name="genomeBuild" type="EntryReference" minOccurs="0"/> </xs:sequence> </xs:choice> <xs:element name="method" type="Method" minOccurs="0" maxOccurs="unbounded" sawsdl:modelReference="http://purl.org/edam/data/0000856"/> <xs:choice maxOccurs="unbounded"> <!--= Annotation by a feature =--> <xs:element name="annotation"> <xs:complexType> <xs:sequence> <!--= Feature type =--> <xs:element name="feature" type="Feature"/> <!--= Feature occurence(s) (instance(s)) =--> <xs:element name="occurence" type="FeatureOccurence" minOccurs="0" maxOccurs="unbounded"/> </xs:sequence> </xs:complexType> </xs:element> <!--= Annotation by a block of inter-related features =--> <xs:element name="blockWithOccurenceReferences"> <xs:complexType> <xs:sequence> <!--= Annotation by a feature (in a block) =--> <xs:element name="annotation" maxOccurs="unbounded"> <xs:complexType> <xs:sequence> <!--= Feature type (in a block) =--> <xs:element name="feature" type="Feature"/> <!--= Feature occurence(s) (instance(s)) (in a block) =--> <xs:element name="occurence" minOccurs="0" maxOccurs="unbounded"> <xs:complexType> <xs:complexContent> <xs:extension base="FeatureOccurence"> <xs:sequence> <xs:element name="association" minOccurs="0" maxOccurs="unbounded"> <xs:complexType> <xs:sequence> <xs:element name="relationName" type="Name" minOccurs="0"/> <xs:element name="relationTerm" type="OntologyTerm" minOccurs="0" maxOccurs="unbounded"/> <xs:element name="associatedFeatureOccurence" type="LocalReference" minOccurs="0" maxOccurs="unbounded"/> </xs:sequence> </xs:complexType> </xs:element> </xs:sequence> <xs:attribute name="localId" type="LocalReference"/> </xs:extension> </xs:complexContent> </xs:complexType> </xs:element> </xs:sequence> </xs:complexType> </xs:element> </xs:sequence> </xs:complexType> <xs:unique name="uniqueLocalId_occ"> <xs:selector xpath="annotation/occurence"/> <xs:field xpath="@localId"/> </xs:unique> </xs:element> </xs:choice> </xs:sequence> </xs:complexType> |
| Model Reference | http://purl.org/edam/data/0001255 |
Evidence complexType
| diagram | 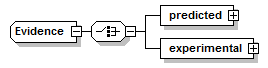 |
||
| children | predicted experimental | ||
| used by |
|
||
| source | <xs:complexType name="Evidence"> <xs:choice> <xs:element name="predicted" type="PredictionResult"/> <xs:element name="experimental" type="ExperimentalEvidence"/> </xs:choice> </xs:complexType> |
MethodResult complexType
| diagram | 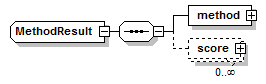 |
||
| children | method score | ||
| used by |
|
||
| source | <xs:complexType name="MethodResult"> <xs:sequence> <xs:element name="method" type="Method"/> <xs:element name="score" type="Score" minOccurs="0" maxOccurs="unbounded"/> </xs:sequence> </xs:complexType> |
Result complexType
| diagram | 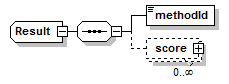 |
||
| children | methodId score | ||
| used by |
|
||
| source | <xs:complexType name="Result"> <xs:sequence> <xs:element name="methodId" type="LocalReference"/> <xs:element name="score" type="Score" minOccurs="0" maxOccurs="unbounded"/> </xs:sequence> </xs:complexType> |
PredictionResult complexType
| diagram | 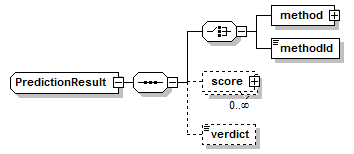 |
||
| children | method methodId score verdict | ||
| used by |
|
||
| source | <xs:complexType name="PredictionResult"> <xs:sequence> <xs:choice> <xs:element name="method" type="Method" sawsdl:modelReference="http://purl.org/edam/data/0000856"/> <xs:element name="methodId" type="LocalReference" sawsdl:modelReference="http://purl.org/edam/data/0000856"/> </xs:choice> <xs:element name="score" type="Score" minOccurs="0" maxOccurs="unbounded"/> <xs:element name="verdict" type="Verdict" minOccurs="0"/> </xs:sequence> </xs:complexType> |
ExperimentalEvidence complexType
| diagram | 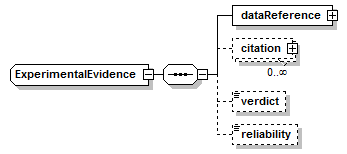 |
||
| children | dataReference citation verdict reliability | ||
| used by |
|
||
| source | <xs:complexType name="ExperimentalEvidence"> <xs:sequence> <xs:element name="dataReference" type="EntryReference" sawsdl:modelReference="http://purl.org/edam/data/0000856"/> <xs:element name="citation" type="EntryReference" minOccurs="0" maxOccurs="unbounded" sawsdl:modelReference="http://purl.org/edam/data/0000970"/> <xs:element name="verdict" type="Verdict" minOccurs="0"/> <xs:element name="reliability" type="Reliability" minOccurs="0"/> </xs:sequence> </xs:complexType> |
Score complexType
| diagram | 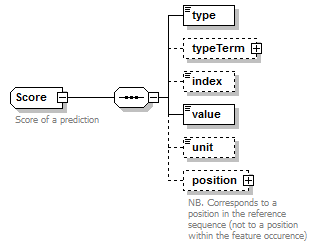 |
||
| children | type typeTerm index value unit position | ||
| used by |
|
||
| Documentation | Score of a prediction | ||
| source | <xs:complexType name="Score" sawsdl:modelReference="http://purl.org/edam/data/0001772"> <xs:annotation> <xs:documentation>Score of a prediction</xs:documentation> </xs:annotation> <xs:sequence> <xs:element name="type" type="ScoreName"/> <xs:element name="typeTerm" type="OntologyTerm" minOccurs="0"/> <xs:element name="index" type="Name" minOccurs="0"/> <xs:element name="value" type="AnyDecimal"/> <xs:element name="unit" type="Name" minOccurs="0"/> <xs:element name="position" type="SequencePosition" minOccurs="0"> <xs:annotation> <xs:documentation>NB. Corresponds to a position in the reference sequence (not to a position within the feature occurence)</xs:documentation> </xs:annotation> </xs:element> </xs:sequence> </xs:complexType> |
||
| Model Reference | http://purl.org/edam/data/0001772 |
GeneralSequencePoint complexType
| diagram | 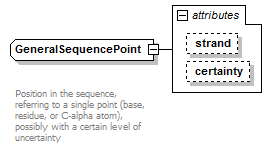 |
||||||||||||||||||
| type | extension of PositiveInteger | ||||||||||||||||||
| properties |
|
||||||||||||||||||
| used by |
|
||||||||||||||||||
| facets |
|
||||||||||||||||||
| attributes |
|
||||||||||||||||||
| Documentation | Position in the sequence, referring to a single point (base, residue, or C-alpha atom), possibly with a certain level of uncertainty | ||||||||||||||||||
| source | <xs:complexType name="GeneralSequencePoint" sawsdl:modelReference="http://purl.org/edam/data/0001016 http://purl.org/edam/format/0002352"> <xs:annotation> <xs:documentation> Position in the sequence, referring to a single point (base, residue, or C-alpha atom), possibly with a certain level of uncertainty </xs:documentation> </xs:annotation> <xs:simpleContent> <xs:extension base="PositiveInteger"> <xs:attribute name="strand" type="Strand"/> <xs:attribute name="certainty" type="Certainty" default="Certain"/> </xs:extension> </xs:simpleContent> </xs:complexType> |
||||||||||||||||||
| Model Reference | http://purl.org/edam/data/0001016 http://purl.org/edam/format/0002352 |
SequencePoint complexType
| diagram | 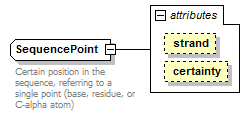 |
||||||||||||||||||
| type | restriction of GeneralSequencePoint | ||||||||||||||||||
| properties |
|
||||||||||||||||||
| used by |
|
||||||||||||||||||
| facets |
|
||||||||||||||||||
| attributes |
|
||||||||||||||||||
| Documentation | Certain position in the sequence, referring to a single point (base, residue, or C-alpha atom) | ||||||||||||||||||
| source | <xs:complexType name="SequencePoint"> <xs:annotation> <xs:documentation>Certain position in the sequence, referring to a single point (base, residue, or C-alpha atom)</xs:documentation> </xs:annotation> <xs:simpleContent> <xs:restriction base="GeneralSequencePoint"> <xs:attribute name="certainty" type="Certainty" fixed="Certain"/> </xs:restriction> </xs:simpleContent> </xs:complexType> |
GeneralSequenceInsertionPoint complexType
| diagram | 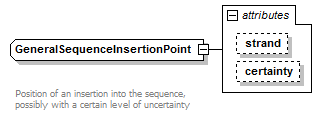 |
||||||||||||||||||
| type | extension of InsertionInteger | ||||||||||||||||||
| properties |
|
||||||||||||||||||
| used by |
|
||||||||||||||||||
| facets |
|
||||||||||||||||||
| attributes |
|
||||||||||||||||||
| Documentation | Position of an insertion into the sequence, possibly with a certain level of uncertainty | ||||||||||||||||||
| source | <xs:complexType name="GeneralSequenceInsertionPoint" sawsdl:modelReference="http://purl.org/edam/data/0001016"> <xs:annotation> <xs:documentation> Position of an insertion into the sequence, possibly with a certain level of uncertainty </xs:documentation> </xs:annotation> <xs:simpleContent> <xs:extension base="InsertionInteger"> <xs:attribute name="strand" type="Strand"/> <xs:attribute name="certainty" type="Certainty" default="Certain"/> </xs:extension> </xs:simpleContent> </xs:complexType> |
||||||||||||||||||
| Model Reference | http://purl.org/edam/data/0001016 |
SequenceInsertionPoint complexType
| diagram | 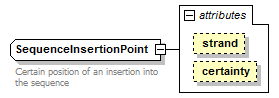 |
||||||||||||||||||
| type | restriction of GeneralSequenceInsertionPoint | ||||||||||||||||||
| properties |
|
||||||||||||||||||
| used by |
|
||||||||||||||||||
| facets |
|
||||||||||||||||||
| attributes |
|
||||||||||||||||||
| Documentation | Certain position of an insertion into the sequence | ||||||||||||||||||
| source | <xs:complexType name="SequenceInsertionPoint"> <xs:annotation> <xs:documentation>Certain position of an insertion into the sequence</xs:documentation> </xs:annotation> <xs:simpleContent> <xs:restriction base="GeneralSequenceInsertionPoint"> <xs:attribute name="certainty" type="Certainty" fixed="Certain"/> </xs:restriction> </xs:simpleContent> </xs:complexType> |
GeneralOutsideSequencePoint complexType
| diagram | 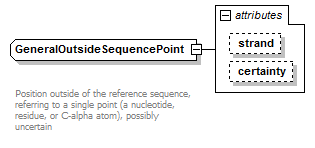 |
||||||||||||||||||
| type | extension of NonzeroInteger | ||||||||||||||||||
| properties |
|
||||||||||||||||||
| used by |
|
||||||||||||||||||
| facets |
|
||||||||||||||||||
| attributes |
|
||||||||||||||||||
| Documentation | Position outside of the reference sequence, referring to a single point (a nucleotide, residue, or C-alpha atom), possibly uncertain | ||||||||||||||||||
| source | <xs:complexType name="GeneralOutsideSequencePoint" sawsdl:modelReference="http://purl.org/edam/data/0001016"> <xs:annotation> <xs:documentation> Position outside of the reference sequence, referring to a single point (a nucleotide, residue, or C-alpha atom), possibly uncertain </xs:documentation> </xs:annotation> <xs:simpleContent> <xs:extension base="NonzeroInteger"> <xs:attribute name="strand" type="Strand"/> <xs:attribute name="certainty" type="Certainty" default="Certain"/> </xs:extension> </xs:simpleContent> </xs:complexType> |
||||||||||||||||||
| Model Reference | http://purl.org/edam/data/0001016 |
OutsideSequencePoint complexType
| diagram | 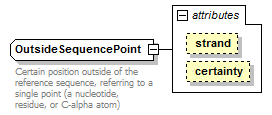 |
||||||||||||||||||
| type | restriction of GeneralOutsideSequencePoint | ||||||||||||||||||
| properties |
|
||||||||||||||||||
| used by |
|
||||||||||||||||||
| facets |
|
||||||||||||||||||
| attributes |
|
||||||||||||||||||
| Documentation | Certain position outside of the reference sequence, referring to a single point (a nucleotide, residue, or C-alpha atom) | ||||||||||||||||||
| source | <xs:complexType name="OutsideSequencePoint"> <xs:annotation> <xs:documentation>Certain position outside of the reference sequence, referring to a single point (a nucleotide, residue, or C-alpha atom)</xs:documentation> </xs:annotation> <xs:simpleContent> <xs:restriction base="GeneralOutsideSequencePoint"> <xs:attribute name="certainty" type="Certainty" fixed="Certain"/> </xs:restriction> </xs:simpleContent> </xs:complexType> |
GeneralSequenceSegment complexType
| diagram | 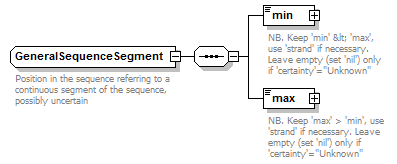 |
||||
| children | min max | ||||
| used by |
|
||||
| Documentation | Position in the sequence referring to a continuous segment of the sequence, possibly uncertain | ||||
| source | <xs:complexType name="GeneralSequenceSegment" sawsdl:modelReference="http://purl.org/edam/data/0001017"> <xs:annotation> <xs:documentation>Position in the sequence referring to a continuous segment of the sequence, possibly uncertain</xs:documentation> </xs:annotation> <xs:sequence> <xs:element name="min" type="GeneralSequencePoint" nillable="true"> <xs:annotation> <xs:documentation>NB. Keep 'min' < 'max', use 'strand' if necessary. Leave empty (set 'nil') only if 'certainty'="Unknown"</xs:documentation> </xs:annotation> </xs:element> <xs:element name="max" type="GeneralSequencePoint" nillable="true"> <xs:annotation> <xs:documentation>NB. Keep 'max' > 'min', use 'strand' if necessary. Leave empty (set 'nil') only if 'certainty'="Unknown"</xs:documentation> </xs:annotation> </xs:element> </xs:sequence> </xs:complexType> |
||||
| Model Reference | http://purl.org/edam/data/0001017 |
SequenceSegment complexType
| diagram | 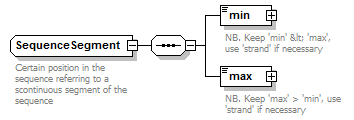 |
||
| type | restriction of GeneralSequenceSegment | ||
| properties |
|
||
| children | min max | ||
| used by |
|
||
| Documentation | Certain position in the sequence referring to a scontinuous segment of the sequence | ||
| source | <xs:complexType name="SequenceSegment"> <xs:annotation> <xs:documentation>Certain position in the sequence referring to a scontinuous segment of the sequence</xs:documentation> </xs:annotation> <xs:complexContent> <xs:restriction base="GeneralSequenceSegment"> <xs:sequence> <xs:element name="min" type="SequencePoint"> <xs:annotation> <xs:documentation>NB. Keep 'min' < 'max', use 'strand' if necessary</xs:documentation> </xs:annotation> </xs:element> <xs:element name="max" type="SequencePoint"> <xs:annotation> <xs:documentation>NB. Keep 'max' > 'min', use 'strand' if necessary</xs:documentation> </xs:annotation> </xs:element> </xs:sequence> </xs:restriction> </xs:complexContent> </xs:complexType> |
GeneralOutsideSequenceSegment complexType
| diagram | 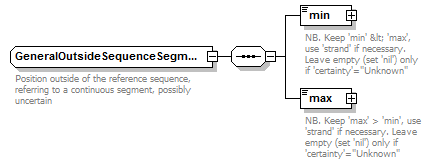 |
||||
| children | min max | ||||
| used by |
|
||||
| Documentation | Position outside of the reference sequence, referring to a continuous segment, possibly uncertain | ||||
| source | <xs:complexType name="GeneralOutsideSequenceSegment" sawsdl:modelReference="http://purl.org/edam/data/0001017"> <xs:annotation> <xs:documentation>Position outside of the reference sequence, referring to a continuous segment, possibly uncertain</xs:documentation> </xs:annotation> <xs:sequence> <xs:element name="min" type="GeneralOutsideSequencePoint" nillable="true"> <xs:annotation> <xs:documentation>NB. Keep 'min' < 'max', use 'strand' if necessary. Leave empty (set 'nil') only if 'certainty'="Unknown"</xs:documentation> </xs:annotation> </xs:element> <xs:element name="max" type="GeneralOutsideSequencePoint" nillable="true"> <xs:annotation> <xs:documentation>NB. Keep 'max' > 'min', use 'strand' if necessary. Leave empty (set 'nil') only if 'certainty'="Unknown"</xs:documentation> </xs:annotation> </xs:element> </xs:sequence> </xs:complexType> |
||||
| Model Reference | http://purl.org/edam/data/0001017 |
OutsideSequenceSegment complexType
| diagram | 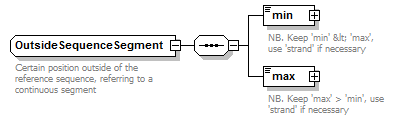 |
||
| type | restriction of GeneralOutsideSequenceSegment | ||
| properties |
|
||
| children | min max | ||
| used by |
|
||
| Documentation | Certain position outside of the reference sequence, referring to a continuous segment | ||
| source | <xs:complexType name="OutsideSequenceSegment"> <xs:annotation> <xs:documentation>Certain position outside of the reference sequence, referring to a continuous segment</xs:documentation> </xs:annotation> <xs:complexContent> <xs:restriction base="GeneralOutsideSequenceSegment"> <xs:sequence> <xs:element name="min" type="OutsideSequencePoint"> <xs:annotation> <xs:documentation>NB. Keep 'min' < 'max', use 'strand' if necessary</xs:documentation> </xs:annotation> </xs:element> <xs:element name="max" type="OutsideSequencePoint"> <xs:annotation> <xs:documentation>NB. Keep 'max' > 'min', use 'strand' if necessary</xs:documentation> </xs:annotation> </xs:element> </xs:sequence> </xs:restriction> </xs:complexContent> </xs:complexType> |
GeneralSequencePosition complexType
| diagram | 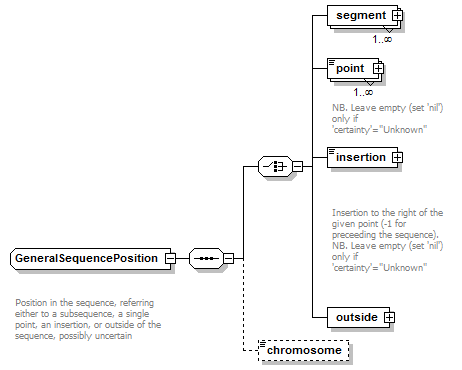 |
||||
| children | segment point insertion outside chromosome | ||||
| used by |
|
||||
| Documentation | Position in the sequence, referring either to a subsequence, a single point, an insertion, or outside of the sequence, possibly uncertain | ||||
| source | <xs:complexType name="GeneralSequencePosition" sawsdl:modelReference="http://purl.org/edam/data/0001014"> <xs:annotation> <xs:documentation> Position in the sequence, referring either to a subsequence, a single point, an insertion, or outside of the sequence, possibly uncertain </xs:documentation> </xs:annotation> <xs:sequence> <xs:choice> <xs:element name="segment" type="GeneralSequenceSegment" maxOccurs="unbounded"/> <xs:element name="point" type="GeneralSequencePoint" nillable="true" maxOccurs="unbounded"> <xs:annotation> <xs:documentation>NB. Leave empty (set 'nil') only if 'certainty'="Unknown"</xs:documentation> </xs:annotation> </xs:element> <xs:element name="insertion" type="GeneralSequenceInsertionPoint" nillable="true"> <xs:annotation> <xs:documentation> Insertion to the right of the given point (-1 for preceeding the sequence). NB. Leave empty (set 'nil') only if 'certainty'="Unknown" </xs:documentation> </xs:annotation> </xs:element> <xs:element name="outside" type="GeneralOutsideSequencePosition"/> </xs:choice> <xs:element name="chromosome" type="ChromosomeName" minOccurs="0"/> </xs:sequence> </xs:complexType> |
||||
| Model Reference | http://purl.org/edam/data/0001014 |
SequencePosition complexType
| diagram | 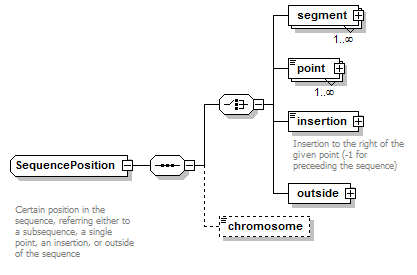 |
||
| type | restriction of GeneralSequencePosition | ||
| properties |
|
||
| children | segment point insertion outside chromosome | ||
| used by |
|
||
| Documentation | Certain position in the sequence, referring either to a subsequence, a single point, an insertion, or outside of the sequence | ||
| source | <xs:complexType name="SequencePosition"> <xs:annotation> <xs:documentation> Certain position in the sequence, referring either to a subsequence, a single point, an insertion, or outside of the sequence </xs:documentation> </xs:annotation> <xs:complexContent> <xs:restriction base="GeneralSequencePosition"> <xs:sequence> <xs:choice> <xs:element name="segment" type="SequenceSegment" maxOccurs="unbounded"/> <xs:element name="point" type="SequencePoint" maxOccurs="unbounded"/> <xs:element name="insertion" type="SequenceInsertionPoint"> <xs:annotation> <xs:documentation>Insertion to the right of the given point (-1 for preceeding the sequence)</xs:documentation> </xs:annotation> </xs:element> <xs:element name="outside" type="OutsideSequencePosition"/> </xs:choice> <xs:element name="chromosome" type="ChromosomeName" minOccurs="0"/> </xs:sequence> </xs:restriction> </xs:complexContent> </xs:complexType> |
GeneralOutsideSequencePosition complexType
| diagram | 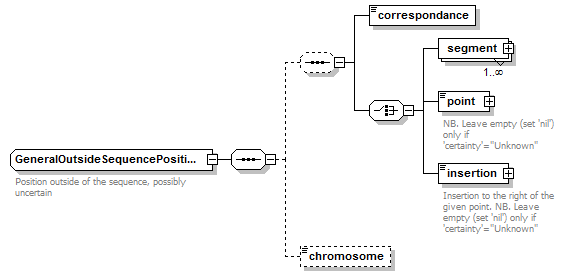 |
||||
| children | correspondance segment point insertion chromosome | ||||
| used by |
|
||||
| Documentation | Position outside of the sequence, possibly uncertain | ||||
| source | <xs:complexType name="GeneralOutsideSequencePosition" sawsdl:modelReference="http://purl.org/edam/data/0001014"> <xs:annotation> <xs:documentation>Position outside of the sequence, possibly uncertain</xs:documentation> </xs:annotation> <xs:sequence> <xs:sequence minOccurs="0"> <xs:element name="correspondance" type="OutsidePositionCorrespondance"/> <xs:choice> <xs:element name="segment" type="GeneralOutsideSequenceSegment" maxOccurs="unbounded"/> <xs:element name="point" type="GeneralOutsideSequencePoint" nillable="true"> <xs:annotation> <xs:documentation>NB. Leave empty (set 'nil') only if 'certainty'="Unknown"</xs:documentation> </xs:annotation> </xs:element> <xs:element name="insertion" type="GeneralOutsideSequencePoint" nillable="true"> <xs:annotation> <xs:documentation>Insertion to the right of the given point. NB. Leave empty (set 'nil') only if 'certainty'="Unknown"</xs:documentation> </xs:annotation> </xs:element> </xs:choice> </xs:sequence> <xs:element name="chromosome" type="ChromosomeName" minOccurs="0"/> </xs:sequence> </xs:complexType> |
||||
| Model Reference | http://purl.org/edam/data/0001014 |
OutsideSequencePosition complexType
| diagram | 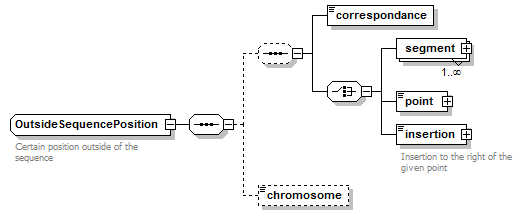 |
||
| type | restriction of GeneralOutsideSequencePosition | ||
| properties |
|
||
| children | correspondance segment point insertion chromosome | ||
| used by |
|
||
| Documentation | Certain position outside of the sequence | ||
| source | <xs:complexType name="OutsideSequencePosition"> <xs:annotation> <xs:documentation>Certain position outside of the sequence</xs:documentation> </xs:annotation> <xs:complexContent> <xs:restriction base="GeneralOutsideSequencePosition"> <xs:sequence> <xs:sequence minOccurs="0"> <xs:element name="correspondance" type="OutsidePositionCorrespondance"/> <xs:choice> <xs:element name="segment" type="OutsideSequenceSegment" maxOccurs="unbounded"/> <xs:element name="point" type="OutsideSequencePoint"/> <xs:element name="insertion" type="OutsideSequencePoint"> <xs:annotation> <xs:documentation>Insertion to the right of the given point</xs:documentation> </xs:annotation> </xs:element> </xs:choice> </xs:sequence> <xs:element name="chromosome" type="ChromosomeName" minOccurs="0"/> </xs:sequence> </xs:restriction> </xs:complexContent> </xs:complexType> |
Gap complexType
| diagram | 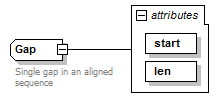 |
||||||||||||||||||
| used by |
|
||||||||||||||||||
| attributes |
|
||||||||||||||||||
| Documentation | Single gap in an aligned sequence | ||||||||||||||||||
| source | <xs:complexType name="Gap"> <xs:annotation> <xs:documentation>Single gap in an aligned sequence</xs:documentation> </xs:annotation> <xs:attribute name="start" type="InsertionInteger" use="required"/> <xs:attribute name="len" type="PositiveInteger" use="required"/> </xs:complexType> |
Frameshift complexType
| diagram | 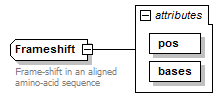 |
||||||||||||||||||
| used by |
|
||||||||||||||||||
| attributes |
|
||||||||||||||||||
| Documentation | Frame-shift in an aligned amino-acid sequence | ||||||||||||||||||
| source | <xs:complexType name="Frameshift"> <xs:annotation> <xs:documentation>Frame-shift in an aligned amino-acid sequence</xs:documentation> </xs:annotation> <xs:attribute name="pos" type="InsertionInteger" use="required"/> <xs:attribute name="bases" use="required"> <xs:simpleType> <xs:restriction base="NonzeroInteger"> <xs:minInclusive value="-2"/> <xs:maxInclusive value="2"/> </xs:restriction> </xs:simpleType> </xs:attribute> </xs:complexType> |
AlignedBiosequence complexType
| diagram | 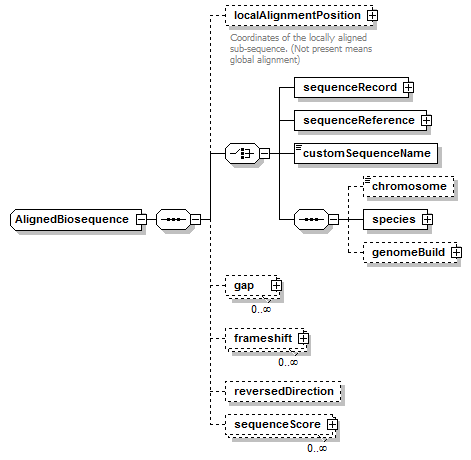 |
||||
| children | localAlignmentPosition sequenceRecord sequenceReference customSequenceName chromosome species genomeBuild gap frameshift reversedDirection sequenceScore | ||||
| used by |
|
||||
| source | <xs:complexType name="AlignedBiosequence"> <xs:sequence> <xs:element name="localAlignmentPosition" type="SequencePosition" minOccurs="0"> <xs:annotation> <xs:documentation>Coordinates of the locally aligned sub-sequence. (Not present means global alignment)</xs:documentation> </xs:annotation> </xs:element> <xs:choice> <xs:element name="sequenceRecord" type="BiosequenceRecord"/> <xs:element name="sequenceReference" type="SequenceReference"/> <xs:element name="customSequenceName" type="Name"/> <xs:sequence> <xs:element name="chromosome" type="ChromosomeName" minOccurs="0"/> <xs:element name="species" type="Species"/> <xs:element name="genomeBuild" type="EntryReference" minOccurs="0"/> </xs:sequence> </xs:choice> <xs:element name="gap" type="Gap" minOccurs="0" maxOccurs="unbounded"/> <xs:element name="frameshift" type="Frameshift" minOccurs="0" maxOccurs="unbounded"/> <xs:element name="reversedDirection" type="Empty" minOccurs="0"/> <xs:element name="sequenceScore" type="Result" minOccurs="0" maxOccurs="unbounded"/> </xs:sequence> </xs:complexType> |
AlignedGeneralNucleotideSequence complexType
| diagram | 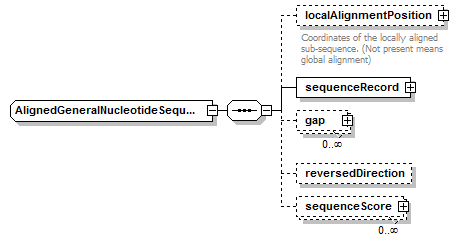 |
||||
| type | restriction of AlignedBiosequence | ||||
| properties |
|
||||
| children | localAlignmentPosition sequenceRecord gap reversedDirection sequenceScore | ||||
| used by |
|
||||
| source | <xs:complexType name="AlignedGeneralNucleotideSequence"> <xs:complexContent> <xs:restriction base="AlignedBiosequence"> <xs:sequence> <xs:element name="localAlignmentPosition" type="SequencePosition" minOccurs="0"> <xs:annotation> <xs:documentation>Coordinates of the locally aligned sub-sequence. (Not present means global alignment)</xs:documentation> </xs:annotation> </xs:element> <xs:element name="sequenceRecord" type="GeneralNucleotideSequenceRecord"/> <xs:element name="gap" type="Gap" minOccurs="0" maxOccurs="unbounded"/> <xs:element name="reversedDirection" type="Empty" minOccurs="0"/> <xs:element name="sequenceScore" type="Result" minOccurs="0" maxOccurs="unbounded"/> </xs:sequence> </xs:restriction> </xs:complexContent> </xs:complexType> |
AlignedGeneralAminoacidSequence complexType
| diagram | 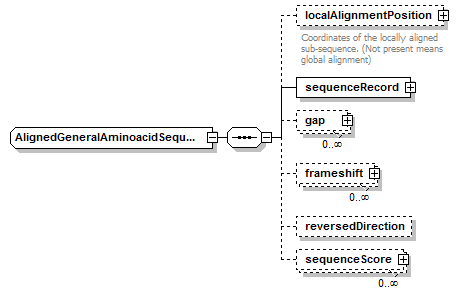 |
||||
| type | restriction of AlignedBiosequence | ||||
| properties |
|
||||
| children | localAlignmentPosition sequenceRecord gap frameshift reversedDirection sequenceScore | ||||
| used by |
|
||||
| source | <xs:complexType name="AlignedGeneralAminoacidSequence"> <xs:complexContent> <xs:restriction base="AlignedBiosequence"> <xs:sequence> <xs:element name="localAlignmentPosition" type="SequencePosition" minOccurs="0"> <xs:annotation> <xs:documentation>Coordinates of the locally aligned sub-sequence. (Not present means global alignment)</xs:documentation> </xs:annotation> </xs:element> <xs:element name="sequenceRecord" type="GeneralAminoacidSequenceRecord"/> <xs:element name="gap" type="Gap" minOccurs="0" maxOccurs="unbounded"/> <xs:element name="frameshift" type="Frameshift" minOccurs="0" maxOccurs="unbounded"/> <xs:element name="reversedDirection" type="Empty" minOccurs="0"/> <xs:element name="sequenceScore" type="Result" minOccurs="0" maxOccurs="unbounded"/> </xs:sequence> </xs:restriction> </xs:complexContent> </xs:complexType> |
AlignedNucleotideSequence complexType
| diagram | 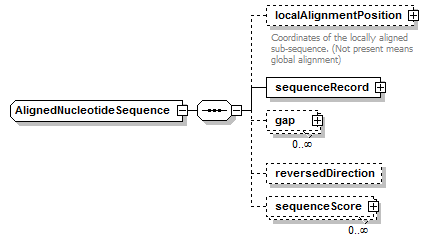 |
||
| type | restriction of AlignedGeneralNucleotideSequence | ||
| properties |
|
||
| children | localAlignmentPosition sequenceRecord gap reversedDirection sequenceScore | ||
| used by |
|
||
| source | <xs:complexType name="AlignedNucleotideSequence"> <xs:complexContent> <xs:restriction base="AlignedGeneralNucleotideSequence"> <xs:sequence> <xs:element name="localAlignmentPosition" type="SequencePosition" minOccurs="0"> <xs:annotation> <xs:documentation>Coordinates of the locally aligned sub-sequence. (Not present means global alignment)</xs:documentation> </xs:annotation> </xs:element> <xs:element name="sequenceRecord" type="NucleotideSequenceRecord"/> <xs:element name="gap" type="Gap" minOccurs="0" maxOccurs="unbounded"/> <xs:element name="reversedDirection" type="Empty" minOccurs="0"/> <xs:element name="sequenceScore" type="Result" minOccurs="0" maxOccurs="unbounded"/> </xs:sequence> </xs:restriction> </xs:complexContent> </xs:complexType> |
AlignedAminoacidSequence complexType
| diagram | 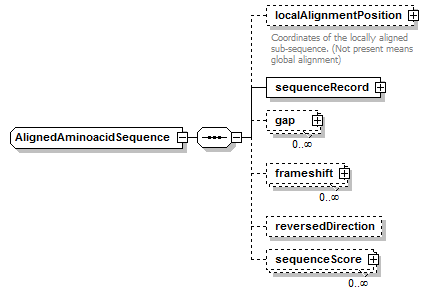 |
||
| type | restriction of AlignedGeneralAminoacidSequence | ||
| properties |
|
||
| children | localAlignmentPosition sequenceRecord gap frameshift reversedDirection sequenceScore | ||
| used by |
|
||
| source | <xs:complexType name="AlignedAminoacidSequence"> <xs:complexContent> <xs:restriction base="AlignedGeneralAminoacidSequence"> <xs:sequence> <xs:element name="localAlignmentPosition" type="SequencePosition" minOccurs="0"> <xs:annotation> <xs:documentation>Coordinates of the locally aligned sub-sequence. (Not present means global alignment)</xs:documentation> </xs:annotation> </xs:element> <xs:element name="sequenceRecord" type="AminoacidSequenceRecord"/> <xs:element name="gap" type="Gap" minOccurs="0" maxOccurs="unbounded"/> <xs:element name="frameshift" type="Frameshift" minOccurs="0" maxOccurs="unbounded"/> <xs:element name="reversedDirection" type="Empty" minOccurs="0"/> <xs:element name="sequenceScore" type="Result" minOccurs="0" maxOccurs="unbounded"/> </xs:sequence> </xs:restriction> </xs:complexContent> </xs:complexType> |
BiosequenceAlignment complexType
| diagram | 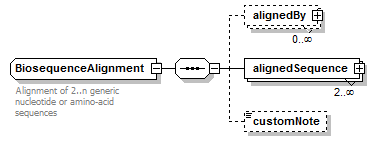 |
||
| children | alignedBy alignedSequence customNote | ||
| used by |
|
||
| Documentation | Alignment of 2..n generic nucleotide or amino-acid sequences | ||
| source | <xs:complexType name="BiosequenceAlignment" sawsdl:modelReference="http://purl.org/edam/data/0000863"> <xs:annotation> <xs:documentation>Alignment of 2..n generic nucleotide or amino-acid sequences</xs:documentation> </xs:annotation> <xs:sequence> <xs:element name="alignedBy" type="MethodResult" minOccurs="0" maxOccurs="unbounded"/> <xs:element name="alignedSequence" type="AlignedBiosequence" minOccurs="2" maxOccurs="unbounded"> <xs:unique name="uniqueGapStart_b"> <xs:selector xpath="gap"/> <xs:field xpath="@start"/> </xs:unique> <xs:unique name="uniqueFrameshiftPos_b"> <xs:selector xpath="frameshift"/> <xs:field xpath="@pos"/> </xs:unique> </xs:element> <xs:element name="customNote" type="FreeText" minOccurs="0"/> </xs:sequence> </xs:complexType> |
||
| Model Reference | http://purl.org/edam/data/0000863 |
GeneralNucleotideSequenceAlignment complexType
| diagram | 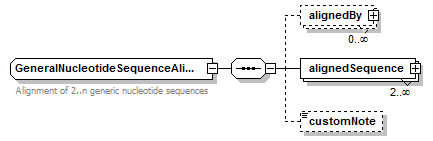 |
||
| type | restriction of BiosequenceAlignment | ||
| properties |
|
||
| children | alignedBy alignedSequence customNote | ||
| used by |
|
||
| Documentation | Alignment of 2..n generic nucleotide sequences | ||
| source | <xs:complexType name="GeneralNucleotideSequenceAlignment" sawsdl:modelReference="http://purl.org/edam/data/0001383"> <xs:annotation> <xs:documentation>Alignment of 2..n generic nucleotide sequences</xs:documentation> </xs:annotation> <xs:complexContent> <xs:restriction base="BiosequenceAlignment"> <xs:sequence> <xs:element name="alignedBy" type="MethodResult" minOccurs="0" maxOccurs="unbounded"/> <xs:element name="alignedSequence" type="AlignedGeneralNucleotideSequence" minOccurs="2" maxOccurs="unbounded"> <xs:unique name="uniqueGapStart_gn"> <xs:selector xpath="gap"/> <xs:field xpath="@start"/> </xs:unique> </xs:element> <xs:element name="customNote" type="FreeText" minOccurs="0"/> </xs:sequence> </xs:restriction> </xs:complexContent> </xs:complexType> |
||
| Model Reference | http://purl.org/edam/data/0001383 |
GeneralAminoacidSequenceAlignment complexType
| diagram | 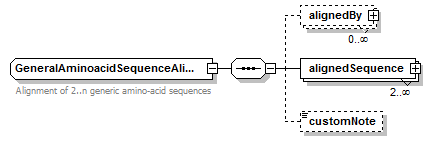 |
||
| type | restriction of BiosequenceAlignment | ||
| properties |
|
||
| children | alignedBy alignedSequence customNote | ||
| used by |
|
||
| Documentation | Alignment of 2..n generic amino-acid sequences | ||
| source | <xs:complexType name="GeneralAminoacidSequenceAlignment" sawsdl:modelReference="http://purl.org/edam/data/0001384"> <xs:annotation> <xs:documentation>Alignment of 2..n generic amino-acid sequences</xs:documentation> </xs:annotation> <xs:complexContent> <xs:restriction base="BiosequenceAlignment"> <xs:sequence> <xs:element name="alignedBy" type="MethodResult" minOccurs="0" maxOccurs="unbounded"/> <xs:element name="alignedSequence" type="AlignedGeneralAminoacidSequence" minOccurs="2" maxOccurs="unbounded"> <xs:unique name="uniqueGapStart_ga"> <xs:selector xpath="gap"/> <xs:field xpath="@start"/> </xs:unique> <xs:unique name="uniqueFrameshiftPos_ga"> <xs:selector xpath="frameshift"/> <xs:field xpath="@pos"/> </xs:unique> </xs:element> <xs:element name="customNote" type="FreeText" minOccurs="0"/> </xs:sequence> </xs:restriction> </xs:complexContent> </xs:complexType> |
||
| Model Reference | http://purl.org/edam/data/0001384 |
NucleotideSequenceAlignment complexType
| diagram | 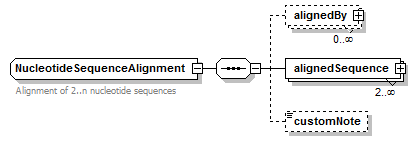 |
||
| type | restriction of GeneralNucleotideSequenceAlignment | ||
| properties |
|
||
| children | alignedBy alignedSequence customNote | ||
| Documentation | Alignment of 2..n nucleotide sequences | ||
| source | <xs:complexType name="NucleotideSequenceAlignment" sawsdl:modelReference="http://purl.org/edam/data/0001383"> <xs:annotation> <xs:documentation>Alignment of 2..n nucleotide sequences</xs:documentation> </xs:annotation> <xs:complexContent> <xs:restriction base="GeneralNucleotideSequenceAlignment"> <xs:sequence> <xs:element name="alignedBy" type="MethodResult" minOccurs="0" maxOccurs="unbounded"/> <xs:element name="alignedSequence" type="AlignedNucleotideSequence" minOccurs="2" maxOccurs="unbounded"> <xs:unique name="uniqueGapStart_n"> <xs:selector xpath="gap"/> <xs:field xpath="@start"/> </xs:unique> </xs:element> <xs:element name="customNote" type="FreeText" minOccurs="0"/> </xs:sequence> </xs:restriction> </xs:complexContent> </xs:complexType> |
||
| Model Reference | http://purl.org/edam/data/0001383 |
AminoacidSequenceAlignment complexType
| diagram | 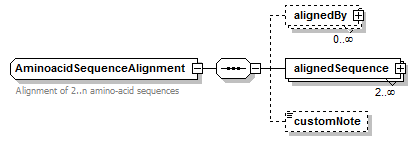 |
||
| type | restriction of GeneralAminoacidSequenceAlignment | ||
| properties |
|
||
| children | alignedBy alignedSequence customNote | ||
| Documentation | Alignment of 2..n amino-acid sequences | ||
| source | <xs:complexType name="AminoacidSequenceAlignment" sawsdl:modelReference="http://purl.org/edam/data/0001384"> <xs:annotation> <xs:documentation>Alignment of 2..n amino-acid sequences</xs:documentation> </xs:annotation> <xs:complexContent> <xs:restriction base="GeneralAminoacidSequenceAlignment"> <xs:sequence> <xs:element name="alignedBy" type="MethodResult" minOccurs="0" maxOccurs="unbounded"/> <xs:element name="alignedSequence" type="AlignedAminoacidSequence" minOccurs="2" maxOccurs="unbounded"> <xs:unique name="uniqueGapStart_a"> <xs:selector xpath="gap"/> <xs:field xpath="@start"/> </xs:unique> <xs:unique name="uniqueFrameshiftPos_a"> <xs:selector xpath="frameshift"/> <xs:field xpath="@pos"/> </xs:unique> </xs:element> <xs:element name="customNote" type="FreeText" minOccurs="0"/> </xs:sequence> </xs:restriction> </xs:complexContent> </xs:complexType> |
||
| Model Reference | http://purl.org/edam/data/0001384 |
RecommendedScoreName simpleType
| type | restriction of Name | ||||||||||||||||||
| used by |
|
||||||||||||||||||
| facets |
|
||||||||||||||||||
| source | <xs:simpleType name="RecommendedScoreName"> <xs:restriction base="Name"> <xs:enumeration value="p-value" sawsdl:modelReference="http://purl.org/edam/data/0001669"/> <xs:enumeration value="E-value" sawsdl:modelReference="http://purl.org/edam/data/0001667"/> <xs:enumeration value="Bit score"/> <xs:enumeration value="Probability"/> </xs:restriction> </xs:simpleType> |
RecommendedVerdict simpleType
| type | restriction of Name | ||||||||||||||||||
| used by |
|
||||||||||||||||||
| facets |
|
||||||||||||||||||
| Documentation | Predefined, recommended verdict of a predicted or experimental evidence | ||||||||||||||||||
| source | <xs:simpleType name="RecommendedVerdict"> <xs:annotation> <xs:documentation>Predefined, recommended verdict of a predicted or experimental evidence</xs:documentation> </xs:annotation> <xs:restriction base="Name"> <xs:enumeration value="Present"/> <xs:enumeration value="Putative"/> <xs:enumeration value="Improbable"/> <xs:enumeration value="Not present"/> </xs:restriction> </xs:simpleType> |
RecommendedReliability simpleType
| type | restriction of Name | ||||||||||||
| used by |
|
||||||||||||
| facets |
|
||||||||||||
| Documentation | Predefined, recommended reliability of an experimental evidence | ||||||||||||
| source | <xs:simpleType name="RecommendedReliability"> <xs:annotation> <xs:documentation>Predefined, recommended reliability of an experimental evidence</xs:documentation> </xs:annotation> <xs:restriction base="Name"> <xs:enumeration value="Reliable"/> <xs:enumeration value="Disputable"/> </xs:restriction> </xs:simpleType> |
QualitativeCertainty simpleType
| type | restriction of Name | |||||||||||||||||||||||
| used by |
|
|||||||||||||||||||||||
| facets |
|
|||||||||||||||||||||||
| Documentation | Qualitative certainty tag | |||||||||||||||||||||||
| source | <xs:simpleType name="QualitativeCertainty"> <xs:annotation> <xs:documentation>Qualitative certainty tag</xs:documentation> </xs:annotation> <xs:restriction base="Name"> <xs:enumeration value="Certain"/> <xs:enumeration value="Uncertain"/> <xs:enumeration value="Unknown"> <xs:annotation> <xs:documentation>The reffered value completely unknown, not the certainty unknown</xs:documentation> </xs:annotation> </xs:enumeration> <xs:enumeration value="Certainly greater or equal"/> <xs:enumeration value="Certainly less or equal"/> </xs:restriction> </xs:simpleType> |
Strand simpleType
| type | restriction of Name | |||||||||||||||||||||
| used by |
|
|||||||||||||||||||||
| facets |
|
|||||||||||||||||||||
| source | <xs:simpleType name="Strand" sawsdl:modelReference="http://purl.org/edam/data/0000853"> <xs:restriction base="Name"> <xs:enumeration value="+"/> <xs:enumeration value="-"/> <xs:enumeration value="+ over start"/> <xs:enumeration value="- over start"/> <xs:enumeration value="Unknown"/> </xs:restriction> </xs:simpleType> |
OutsidePositionCorrespondance simpleType
| type | restriction of Name | |||||||||||||||||||||||||||||||
| used by |
|
|||||||||||||||||||||||||||||||
| facets |
|
|||||||||||||||||||||||||||||||
| Documentation | Correspondance of the outside positions to the reference sequence | |||||||||||||||||||||||||||||||
| source | <xs:simpleType name="OutsidePositionCorrespondance"> <xs:annotation> <xs:documentation>Correspondance of the outside positions to the reference sequence</xs:documentation> </xs:annotation> <xs:restriction base="Name"> <xs:enumeration value="In chromosome"> <xs:annotation> <xs:documentation> The outside position is in the nucleotide sequence of the either explicitly given chromosome or the chromosome of the reference sequence. Outside-positions 1..m are positions in the chromosome </xs:documentation> </xs:annotation> </xs:enumeration> <xs:enumeration value="In referenced supersequence"> <xs:annotation> <xs:documentation> The outside position is in an explicitly referenced nucleotide or amino-acid supersequence (respectively) of the reference sequence. Outside-positions 1..m are in the explicitly referenced supersequence </xs:documentation> </xs:annotation> </xs:enumeration> <xs:enumeration value="In implicit supersequence"> <xs:annotation> <xs:documentation> The outside position is in a nucleotide or amino-acid supersequence (respectively) of the reference subsequence. Positions 1..n correspond, position -1 is the preceding (next point to the left) from 1 </xs:documentation> </xs:annotation> </xs:enumeration> <xs:enumeration value="Relative to 1st translated base"> <xs:annotation> <xs:documentation> The outside position is in a nucleotide sequence. Position 1 corresponds to the 1st base of the 1st translated codon within the reference isoform, position -1 is the next to the left from 1 (NB. CDSs include the start and stop codons: do not use this correspondance option for outside features of CDSs) </xs:documentation> </xs:annotation> </xs:enumeration> <xs:enumeration value="Relative to 1st transcribed base"> <xs:annotation> <xs:documentation> The outside position is in a nucleotide sequence. Position 1 corresponds to the 1st base transcribed within the reference isoform, position -1 is the next to the left from 1 </xs:documentation> </xs:annotation> </xs:enumeration> </xs:restriction> </xs:simpleType> |
RecommendedDatabaseName simpleType
| type | restriction of Name | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| used by |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| facets |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Documentation | Predefined, recommended database name of a public database | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| source | <xs:simpleType name="RecommendedDatabaseName"> <xs:annotation> <xs:documentation>Predefined, recommended database name of a public database</xs:documentation> </xs:annotation> <xs:restriction base="Name"> <xs:enumeration value="EMBL"/> <xs:enumeration value="EMBLCON"/> <xs:enumeration value="EMBLANN"/> <xs:enumeration value="EMBLCDS"/> <xs:enumeration value="EMBLSVA"/> <xs:enumeration value="Ensembl Gene"/> <xs:enumeration value="Ensembl Transcript"/> <xs:enumeration value="UniProt" sawsdl:modelReference="http://purl.org/edam/resource/0000651"/> <xs:enumeration value="UniProt/Swiss-Prot" sawsdl:modelReference="http://purl.org/edam/resource/0000649"/> <xs:enumeration value="UniProt/TrEMBL" sawsdl:modelReference="http://purl.org/edam/resource/0000650"/> <xs:enumeration value="UniSave"/> <xs:enumeration value="UniRef100"/> <xs:enumeration value="UniRef90"/> <xs:enumeration value="UniRef50"/> <xs:enumeration value="UniParc"/> <xs:enumeration value="IPI"/> <xs:enumeration value="RefSeq"/> <xs:enumeration value="InterPro"/> <xs:enumeration value="PDB"/> <xs:enumeration value="HGVbase"/> <xs:enumeration value="GenomeReviews"/> <xs:enumeration value="EPO Proteins"/> <xs:enumeration value="JPO Proteins"/> <xs:enumeration value="KIPO Proteins"/> <xs:enumeration value="USPO Proteins"/> <xs:enumeration value="Medline"/> <xs:enumeration value="DDBJ/GenBank/EMBL"/> <xs:enumeration value="UniProt"/> <xs:enumeration value="PDB"/> <xs:enumeration value="DAD"/> <xs:enumeration value="PRF"/> <xs:enumeration value="Patent"/> <xs:enumeration value="NCBI Taxonomy" sawsdl:modelReference="http://purl.org/edam/resource/0000833"/> <xs:enumeration value="NCBI Genetic Code"/> </xs:restriction> </xs:simpleType> |
RecommendedOntologyName simpleType
| type | restriction of Name | |||||||||||||||
| used by |
|
|||||||||||||||
| facets |
|
|||||||||||||||
| Documentation | Predefined, recommended ontology name of a public ontology | |||||||||||||||
| source | <xs:simpleType name="RecommendedOntologyName"> <xs:annotation> <xs:documentation>Predefined, recommended ontology name of a public ontology</xs:documentation> </xs:annotation> <xs:restriction base="Name"> <xs:enumeration value="Gene Ontology" sawsdl:modelReference="http://purl.org/edam/resource/0000830"/> <xs:enumeration value="Sequence Ontology"/> <xs:enumeration value="Protein Features Ontology"/> </xs:restriction> </xs:simpleType> |